
Hello, I’m Ryan.
I’m a Biology major and plan to attend Eastern Virginia Medical School for Master in Physician Assistant Studies once I graduate. I grew up in Virginia Beach and moved to Norfolk in 2017. My major hobbies are surfing and adventuring with my fiancée and the dogs.
Introduction to Immunology
-Mab Drug:
Ansuvimab is a monoclonal antibody drug used to treat humans infected with Zaire ebolavirus, more commonly known as Ebola. Ebola is a single-stranded RNA virus with an extremely high fatality rate and was discovered in 1976. There are five species, with Zaire ebolavirus (ZEBOV) and Sudan ebolavirus (SEBOV) being responsible for most outbreaks. Ebola viruses belong to the Filoviridae family whose viral characteristics include the following: enveloped, non-segmented RNA genome, and filamentous particles. The Ebola virus consists of seven genes that code for nucleoprotein, glycoprotein, RNA-dependent RNA polymerase (L)-5’ trailer, and the virion proteins VP35, VP40, VP30, and VP24. The viral genome is protected by a nucleocapsid made up of VP30 and nucleoprotein. VP35 and RNAdependent RNA polymerase help form the replication complex and VP24 and VP40 are matrix proteins involved in particle formation. VP24 and VP35 can also act as interferon antagonists (Reece et al., 2016).
Ebola deaths are due to systemic shock from vascular dysfunction. Vascular permeability and disseminated intravascular coagulation increase and immunological responses are impaired in infected individuals. The virus initially infects monocytes, macrophages, and dendritic cells which result in the activation and reduction in barrier function of endothelial cells. Infected monocytes secrete proinflammatory factors IL-6, IL-1beta, TNF-alpha, and nitric oxide; the latter two are believed to be responsible for the decrease in endothelial barrier function. The virus replicates rapidly and may induce expression of tissue factor which can downregulate thrombomodulin resulting in coagulopathy. This clotting impairment often fuels inflammation which further induces coagulopathy, which further induces inflammation, and so on. This cycle is part of the system shock that kills so many infected persons (Brown et al., 2008).
Ansuvimab is an Immunoglobulin G antibody that was isolated from an Ebola survivor’s blood in 1995. It showed promising neutralization of the virus, and the medication was approved for use in the United States in December of 2020. Ansuvimab works by disrupting a crucial step in Ebola infections: the binding of the viral glycoprotein to the Niemann-Pick C1 (NPC1) protein (Lee, 2021). NPC1 is a lateendosomal membrane protein involved in transporting LDL-derived cholesterol (Li et al., 2016). Ansuvimab binds the LEIKKPDGS epitope of the receptor binding site in the GP1 subunit of the glycoprotein. Ansuvimab has been shown by biolayer interferometry to have a high affinity for GP1 and to have an inhibitory effect on the binding of GP1 to NPC1 (Lee, 2021). In simpler terms, the medicine inhibits viral glycoproteins and viral internalization by cells. Patient survival is increased by a disruption
in the viral life cycle by Ansuvimab.
End-of-term reflection:
Introduction to Immunology is certainly among the most difficult courses I’ve had in my undergraduate studies. I think learning about MHC I helped me better understand how the immune system functions, and more specifically the activity of NK cells. I remember learning in A&P that natural killer cells kind of circulate looking for non-self cells to kill. That wasn’t wrong, but I didn’t get the full picture. Now I understand that NK cells specifically target cells lacking MHC I; I also know some of their killing mechanisms. Still, it’s more complicated than just MHC: NK cells have a variety of receptors that either inhibit or promote cytotoxic activity. Cells expressing MHC I, but otherwise altered into an unhealthy state, may still be targeted by NK cells. NK cells, kind of like cytotoxic T cells, release granzyme and perforin to trigger apoptosis in their targets. Fascinating.
Before learning some immunology, I knew cell signaling was complex but I had no idea just how intricate immune cells were. For example, the ability of B cells to have nearly infinite receptor diversity through recombination kind of blew my mind. In January I thought the immunology content was going to be easy, but it turned out to be a bit overwhelming. But sitting at the end of the semester, I’ve realized how much I soaked in. Throughout my undergrad, I’ve developed somewhat of a habit of skimming biology articles to fulfill curiosity. Now I feel I can understand even more with my newfound immunology foundation.
Sugar-Free Challenge (Strict)
I was excited to do the sugar-free challenge because I thought it would be sort of a good reset. Historically, I did not consume excessive added sugars because most of my meals are made up of fresh foods, so I wasn’t concerned about how difficult the challenge would be. I also thought after the challenge I would feel so good that I would keep my added sugar intake to a minimum. This was not the case!
Day one – Mon 10/24:
Breakfast
Black coffee
Oats, walnuts, banana, cinnamon
Midday
Smoothie with oats, 2 bananas, blueberries, spinach and water
1 apple
Dinner
Sheet-pan rice and veggies with tofu
This is one of my favorite meals because it’s so easy and versatile. I eat this at least once a week.
Recipe:
8 oz tofu
1 cup rice
2-3 cups veggie stock (depending on white or brown rice)
Small head of broccoli
¾ red pepper
4 large mushrooms
(just about any mixture of veggies works)
Cover with aluminum foil and bake at 350
It’s good to make a sauce and add it when rice is almost done and cook for 5 or 10 more minutes
I don’t remember the measurements I just eyeball it:
soy sauce, balsamic vinegar, sesame oil, chili flakes, corn starch, fresh garlic, fresh ginger
2 pears
This day was pretty typical as far as my diet goes in general. I noticed a couple differences, however. I drank less coffee; I think because normally I add sugar which makes it much more appealing. I’ll usually go back for a second or third cup before leaving the house. Most days I wash down my breakfast with a small glass of orange juice as well which was only slightly disappointing to not have. I felt great throughout the day though a bit sleepy in the afternoon. After dinner was when I noticed something was missing. I’m a big fan of eating sweets after dinner. My most frequent choices usually involve chocolate, ice cream, or some combination of the two. Chocolate covered nuts are probably my favorite sweet nighttime snack. There was plenty of chocolate in the house, and someone was there that had no interest in the sugar-free challenge eating chocolate in front of me! I ended up eating two pears which didn’t really scratch the itch but seemed to do something. I went to sleep feeling a bit full but good, nonetheless.
Day two – Tues 10/25:
Breakfast
Black coffee
Scrambled eggs with cheese, two slices avocado toast
Midday
Smoothie with oats, 2 bananas, blueberries, spinach, peanut butter and water
1 pear
Like 30 pistachios
Dinner
Shrimp fajitas (minus the wraps, I couldn’t find any w/o added sugar)
Apple and peanut butter for dessert
I felt great in the morning and had a nice home workout. I was disappointed because I couldn’t add protein powder to my smoothie as it was loaded with added sugar. Instead, I added peanut butter. Like Monday morning I did not have as much coffee. It started to not be my favorite day. I began to feel irritable around 10:30 and a bit foggy headed. I don’t know if I can attribute that to the lack of added sugars because sometimes you just have those days. Still, I have my suspicions because I usually feel sharp and more stoic on workout days. It could have been the slight reduction in caffein as well. The day was uneventful, but some small things were annoying me, and I just felt a little mad at the world. The after-dinner sugar cravings hit me again and the apple with peanut butter didn’t satisfy. Chocolate was calling my name. I also had a long stare down with the orange juice but stayed strong and hit the hay.
Day three – Wed 10/26:
Breakfast
2 cups black coffee
Scrambled eggs with cheese, two slices avocado toast (Ezekiel bread)
Midday
Smoothie with oats, 2 bananas, blueberries, spinach and water
Leftover rice/veggies
Dinner
Cabbage and potato soup
I felt a bit blah in the morning and irritable until the afternoon although I had 2 cups of coffee. So, I think my body was still adjusting to no sugar in the morning. Overall, the day was good, and I didn’t feel any remarkable way. That night however, I had insane sugar cravings after dinner and ended up eating 4 pears back-to-back and I still wanted more. I was very stuffed though, so I had to stop. The pears were so ripe and delicious but for some reason fruit doesn’t make me happy like chocolate and ice cream do.
Day four – Thurs 10/27:
Breakfast
Black coffee
Scrambled eggs with cheese, two slices avocado toast
Midday
Smoothie with oats, 2 bananas, blueberries, spinach, peanut butter and water
Dinner
Left over soup and toast
By day four I felt good. My coffee intake was probably a little more than half what I would usually drink by the fourth day of the week. I was no longer irritable or feeling blah and had a great workout before leaving the house. I probably would have gone crazy by now if it wasn’t for the daily smoothies loaded with fructose. I did have another stare-down Thursday evening though, this time with a big bag of Halloween candy. Once again, the post-dinner sugar cravings hit me but I was out of pears and wasn’t in the mood for an apple.
Day five – Fri 10/28:
Breakfast
Black coffee
Scrambled eggs with cheese, two slices avocado toast
Midday
Smoothie with oats, 2 bananas, blueberries, spinach and water
Dinner
Homemade pizza
I felt good on the last day. I think I’d fully adjusted by Friday. I was ready for the challenge to be over though. There was a point Friday morning where I wanted to snack on some trail mix but it had some little chocolate candies in it and I was disappointed. Technically the first sweet I had after the sugar-free challenge was sugar in my coffee. It was glorious. Though I didn’t use as much sugar as normal, I drank three small cups. No regrets. That night was the real reward. CHOCOLATE. I ate a quarter of a fancy dark chocolate bar and 2 bite size twix. The twix were not as good as I expected, maybe it’s me or the sugar-free challenge, but the milk chocolate didn’t taste all that good. I was still happy to be free to satisfy my nighttime cravings though. A few days out from the challenge and I was consuming added sugars like nothing ever happened. I was able to resist the Halloween candy binge though. I think the challenge strengthened my resistance to impulse sweet eating. Sunday I ate a breakfast burrito, which I missed so much during the challenge because the wraps always have added at least 1g sugar.
Genetics
Writing Assignment # 1 – Personal Statement
After graduating high school, I had no clue what I wanted to do for a living. One day when driving around Virginia Beach some insight came my way. I saw a car accident that must have occurred just minutes before I arrived as there were no first responders in sight. I pulled over to see if I could help and saw an SUV bottom-side up with a woman trapped in the driver’s seat. She had a small stream of blood on her face and was consoling her daughter in the back seat. Moments later police arrived and minutes later firefighters. By this time there were about a dozen other bystanders ready to lend a hand. After evaluating the situation, the firefighters beckoned us to come and help lift the SUV so they could pull the two occupants out. It took maybe a minute or two for us to lift the SUV slightly and for the firefighters to free them. Once we set the vehicle back down, I went on my way feeling proud and exhilarated. I thought to myself “that’s something I could do, something I would love to do. Helping people when they’re truly in need.” Despite that experience, I was still somewhat lost. After working various jobs in the seven or eight years between graduating high school and beginning my college education, I realized that I wanted to help people in a more meaningful way. Yes, I was helping people when I was flipping burgers, when I was a laborer renovating houses, when I operated carnival games, and when I was a supervisor at the busiest Wawa in Hampton Roads. The problem was that I didn’t feel fulfilled. I felt more like a cog in a machine of another’s design than an independent contributor of goodness to the world. Thinking back to the trapped mother and daughter and the heroism I felt that day, I knew my vocation would involve saving lives.
Since I began my higher education in 2017, I’ve maintained the dream of attending
graduate school and ultimately a career as a physician assistant. Some people ask why I am
choosing a PA program versus the medical doctor path and there are a few reasons. One reason
has to do with my personality. I’ve noticed that the feeling of excitement and fascination for
some novel activity, be it athletic, occupational or leisure, tends to quickly fade for me.
Considering this trait, an MD program is less appealing due to the fear of being in too deep
before realizing the specialty I choses is no longer so interesting. The PA profession however
generally allows one to change specialties more easily. I also chose the PA route because I
didn’t begin higher education until I was 27. I’m 32 now, will be married soon, and the idea of a
family is on the horizon. I would like to be established in my career within four years and ready
to begin the next great journey in life: raising children. Furthermore, the physician assistant
training would check two boxes for me; provide a fulfilling career, and the knowledge I need to
be helpful, or not helpless, in a medical emergency.
After starting college, I reached the next milestone when I was hired as a medical
assistant in March of 2018. I remember having no real medical knowledge at the time except
maybe RICE (rest, ice, compression, elevation). The training and experience I’ve gained since
have been invaluable. From measuring blood pressure, to venipuncture, basic life support,
performing EKG’s, preparing and applying fiberglass splints, consulting providers, properly
triaging patients, understanding HIPAA compliance; the list goes on. I’m constantly growing,
and my resolve hasn’t wavered. Five years later and I can feel the culmination of my efforts
bringing me closer to medical school. I want to be a person with the knowledge and experience to help those in need. I want to be a healthcare provider. I’m grateful for your consideration and
look further to an interview in which I can address any queries and further express my ambitions.
Writing Assignment # 2
A primary article is one which reports novel research findings. Primary articles are usually peerreviewed and divided into the sections Introduction, Methods, Results, Discussion, and References. It’s
important to note that primary articles are not a summary of pre-published information and prior findings
but rather a contribution of new information to the literature.
Review articles on the other hand are a synthesis of information that has already been published
on the topic. They are a compilation, summary, and interpretation of past articles and therefore
considered a secondary source. These sorts of articles can be particularly useful when learning new
material.
The scientific peer-review process is in place to screen articles for significance and quality.
Articles are only approved if they have proper methodology, original findings, and information of
significance relative to the field of study. Scientific articles are written for an audience of other
researchers; hence the term “peer” in peer-review. Once an article is submitted for publication to a
scholarly journal, the editor determines if the article is a good candidate. If the article makes it passed the
editor, it’s then reviewed by a panel of experts, made up of the author’s peers, who evaluate the article’s
quality and significance and provide their recommendation back to the editor. Scientific journals often
have high rejection rates, and the peer-review process can be quite lengthy. It can take months or years,
and possibly many revisions, before the article reaches journal publication. The journal editor makes the
final decision to feature the article or not. The peer-review process is still being optimized as scientists
aren’t always in consensus regarding peer-review methodology.
The article Huntington’s Disease: Mechanisms of Pathogenesis and Therapeutic Strategies by
Jimenez-Sanchez et al. is an example of a review article. This is apparent in the introductory section in
which the authors explicitly state the paper is a review and discussion of the nature of Huntington’s
Disease, and an outline of the then-current therapies. The article Permanent inactivation of Huntington’s
disease mutation by personalized allele-specific CRISPR/Cas9 by Shin et al. is a primary article. One can
tell the article is primary because it doesn’t only describe known information on Huntington’s disease,
rather the article adds information, derived by the authors themselves, to the sphere of knowledge. The
authors performed a study, which had not been conducted before, that yielded novel findings.
Writing Assignment # 3 – Primary Article Citation
Gazestani, Vahid H. et al. A perturbed gene network containing PI3K-AKT, RAS-ERK, and WNT-ß-catenin pathways in leukocytes is linked to ASD genetics and symptom severity. Nature neuroscience22, 1624-1634 (2019). https://doi.org/10.1038/s41593-019-0489-x
Writing Assignment # 4 – Article Summary
The article published in Nature Neuroscience in 2019 ‘A perturbed gene network containing
PI3K-AKT, RAS-ERK and WNT-β-catenin pathways in leukocytes is linked to ASD genetic and
symptom severity’ by Gazestani et al. shed some light on the genetics of prenatal and early postnatal brain development in toddlers with autism spectrum disorder (ASD). By analyzing the leukocyte
transcriptomes of 1-to-4 year-old male toddlers with typical development (TD) or ASD, the authors
discovered a dysregulated gene network that includes genes highly expressed during fetal brain
development.
The authors found a differentially expressed (DE) ASD network which had stronger co-
expression in toddlers with ASD than those with TD. Lists of ASD risk (rASD) genes were extracted
from the SFARI database and from prior data gathered and compared with the DE-ASD network which
showed significant overlap with regulatory targets of rASD genes. The authors also identified key
pathways involved in expanded ASD (XP-ASD) and DE-ASD networks and demonstrated by enrichment
analysis that the XP-ASD network is highly enriched for signaling pathways. DE genes were more
enriched for cell proliferative processes when compared to rASD genes in the XP-ASD network,
especially PI3K-AKT and associated pathways. However, rASD genes showed to be more enriched for
neuron differentiation and maturation. By scoring gene mutations based on their effects on the
phosphorylation state of ERK, AKT, and β-catenin proteins, rASD genes in the XP-ASD network were
notably enriched for regulators of RAS-ERK, PI3K-AKT, and WNT-β-catenin pathways. However, no
significant enrichment for regulators of the same pathways were observed among rASD genes that
weren’t included in the XP-ASD network. The results supported the idea that rASD genes regulate the
DE-ASD network by perturbing the RAS-ERK, PI3K-AKT, and WNT-β-catenin pathways. To validate
the association of increased coexpression in the DE-ASD network with neural proliferation and
maturation processes, the DE-ASD networks of toddlers with ASD were examined to determine if the
DE-ASD network showed increased coexpression in human induced pluripotent stem cell (hiPSC)-
derived progenitors and neurons. The analysis demonstrated the DE-ASD network is indeed more active
in the neural models of people with ASD which was further supported by the presence of macrocephaly, a
common phenotype in ASD, in the ASD individuals studied. The toddler’s ASD symptom severity was
also evaluated to test whether the magnitude of coexpression activity of the DE-ASD network correlated
with Autism Diagnostic Observation Schedule social affect (ADOS-SA) scores. The authors confirmed a
correlation between the extent of gene coexpression activity in the DE-ASD network and ADOS-SA
scores, and therefore ASD severity.
It’s known that ASD has a significant genetic basis, but how rASD genes relate to molecular
variations which contribute to ASD is poorly understood. The article however illuminated a dysregulated
gene network that is strongly associated with high confidence rASD genes and provided evidence of
shared mechanisms underlying ASD. The authors propose that further studies with larger datasets should
be performed both to replicate their results but also to tackle other questions on the matter such as the sex bias seen in ASD or molecular mechanisms differentiating low and high-functioning individuals.
Writing Assignment #5
The Virginian-Pilot article ‘Simple blood test could detect cancer early – Scientists
consider easy, cheap ‘liquid biopsy’ screening’ by author Lisa Krieger highlights potential broad
cancer screenings that may soon be widely available due to recent advances in genetics and
bioinformatics. By testing one’s blood for various cancer biomarkers, cancer could be detected
well before symptoms arise. However, at this stage the specificity may not be sharp enough to
improve patient outcomes and may instead induce psychological stress or even be a false alarm.
‘Liquid biopsy’ uses the word biopsy, usually attributed to a cell or tissue sample, that is
collected then observed under a microscope, or otherwise evaluated for malignancy. In this case,
the tissue is blood, which is considered a connective tissue in human biology. Cancer
biomarkers include a variety of biomolecules, and historically cancer screening has only been
available for breast, cervical, colorectal, lung, and prostate cancer; each screening only tests for
an individual cancer type. The author goes on to discuss a field called multi-cancer early
detection (MCED), which aims to integrate liquid biopsies into standard medical practice. The
logic behind the multi-cancer screening is that as cells age and perish, their genetic material is
shed into the bloodstream, with cancer cells being no exception.
The major technological innovations contributing to more sophisticated cancer screening
methods are in genomics and bioinformatics. High throughput gene sequencing, according to
Dr. Maximilian Diehn with Stanford Medicine oncology, can sequence millions and billions of
DNA molecules in one blood sample in search for mutant molecules. Modern bioinformatics
can analyze the incredibly numerous data points for specific markers such as DNA methylation,
extra or missing chromosomes, and cancer suggestive RNAs and proteins.
Several companies and research labs are developing these sorts of tests including GRAIL,
a biotech company which currently has a $949 test on the market for prescribed patients over 50
or otherwise at a greater risk of cancer. Still, the tests are in the early stages, and it’s unclear
how patient outcomes will be improved by such tests. Some experts are concerned anxieties
from the results and further testing could be detrimental to patients. Also, there are concerns
about the tests overrepresenting cancer biomarkers and about taxpayers suffering due to a new,
expensive test offered annually to Medicare beneficiaries.
I believe the information in the article is accurate and is supported by a Cancer Cell
article titled ‘New genomic technologies for multi-cancer early detection: Rethinking the scope
of cancer screening’ by Hackshaw, Clarke, and Hartman. The Virginian-Pilot article briefly
summarized the topic and highlighted the current limitations of the technology well. MCED
tests targeting numerous cancer types vary in sensitivity, with the sensitivity decreasing as the
number of cancer types studied increase. False positive rates also vary but remain between 0-4%.
The major benefit of MCEDs seems to be in screening for cancers which currently have no
effective tests. These cancers account for 70-75% of all cancer deaths. Yet, it is doubtful how
screening for these cancers could ever be cost effective on the population scale. Ultimately,
MCED tests are expected to reduce cancer morbidity and mortality in the future but at the current stage it is unclear how healthcare providers would implement MCED testing to large populations
at an affordable rate.
Sources:
Hackshaw, Allen, Clarke, Christina A., Hartman, Anne-Renee. New genomic technologies for
multi-cancer early detection: Rethinking the scope of cancer screening. Cancer Cell 40, 109-113
(2022) https://doi.org/10.1016/j.ccell.2022.01.012
Krieger, Lisa M.. Simple blood test could detect cancer early – Scientists consider easy, cheap
‘liquid biopsy’ screenings. The Virginian-Pilot, 9D (2023).
Romanov Assignment – A
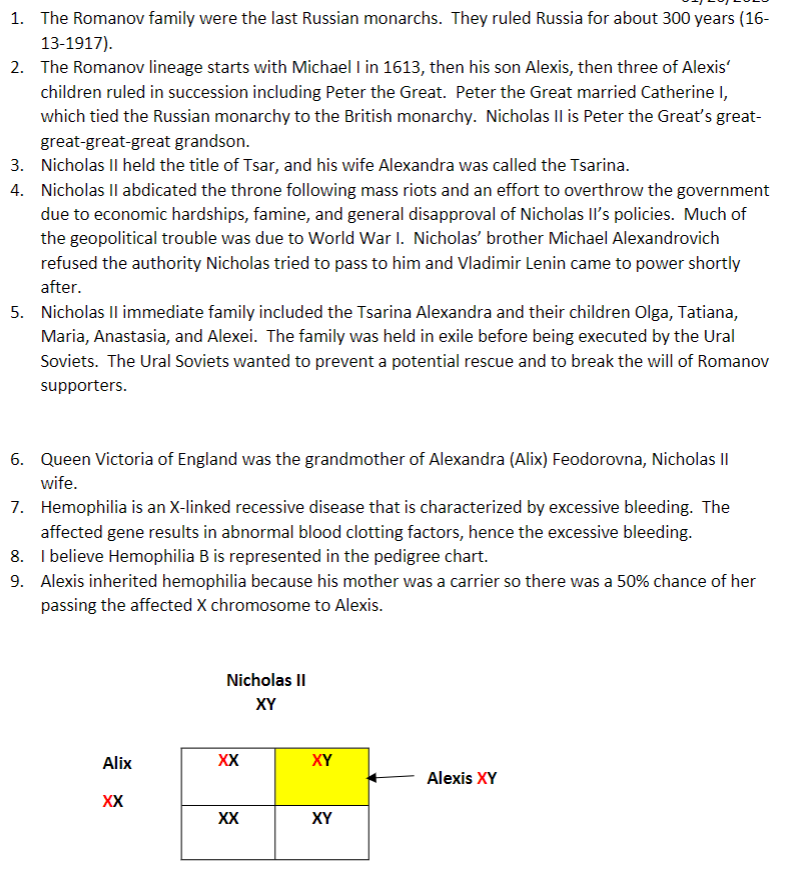
1. Who were the Romanov’s (in Russian history)?
2. Briefly outline the Romanov lineage.
3. Nicholas II was the last Romanov to hold power in Russia. What was his title?
4. What happened to Nicholas II? Why (from a geopolitical view)? Who then took control?
5. Describe the family of Nicholas II. What happened to them?
6. How was Nicholas II wife, Alix, related to Queen Victoria of England? Both Queen Victoria and Alix are designated as being carriers for hemophilia.
7. In a couple of sentences, describe the disease hemophilia.
8. What type of hemophilia (A or B) is (probably) represented in the pedigree chart?
9. The Romanov’s son, Alexis, had hemophilia. Describe how Alexis genetically acquired hemophilia.
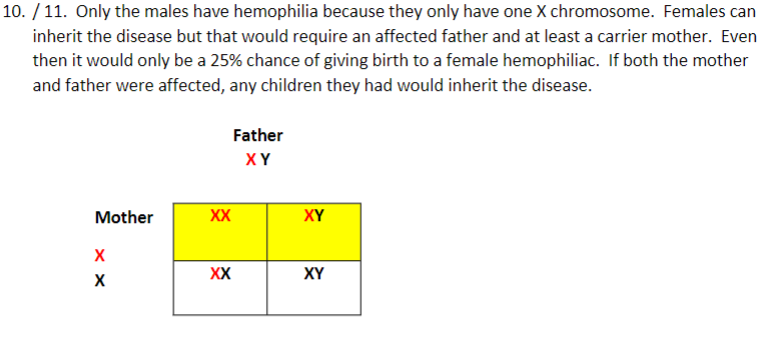
10. Using a Punnett square (again, draw a table or line up the genotypes), explain why only males in the pedigree chart have hemophilia.
11. Is it possible for a female to inherit hemophilia, and, if so, how?
12. None of Alexis’ sisters are shown to have hemophilia. Using only the tools available at the time they lived, how could it have been determined whether they were carriers like their mother.
13. Using a Punnett square (again, draw a table or line up the genotypes), what is the probability the daughter of a mother who is a carrier and a father who does not have the disease, will be a carrier?
14. Using a Punnett square (again, draw a table or line up the genotypes), what is the probability that 4 daughters of a mother who is a carrier and a father who does not have the disease, will be a carrier?
15. Using a Punnett square (again, draw a table or line up the genotypes), explain why none of Alexi’s sisters had hemophilia.
Romanov Assignment – B
1. Using your knowledge from Module 4, on what chromosome is the gene that causes hemophilia?
2. Describe the mutation that apparently caused hemophilia in Alix, (and probably all of the European families that had hemophilia).
3. Using your knowledge from Module 7, describe how the mutation you described in #10 could result in a faulty gene product.
4. Again, using your knowledge from Module 4, give the genotype for a carrier of hemophilia.
5. Mitochondrial DNA testing was also done on both Nicholas II and Alix. Why was information from Alix’s, but not Nicholas’, mitochondrial DNA used to identify three females as belonging to Alix?
6. HRH Prince Philip, the Duke of Edinburgh, provided mitochondrial DNA used to identify Alix and her three daughters. Why was his mitochondrial DNA used?
Who was the HRH Prince Philip, the Duke of Edinburgh in today’s world? Do you ever hear of his grandchildren (in magazines while you are waiting to check out of a store)?
7. Who was missing from the mass grave?
8. The Duke of Fife and Princess Xenia provided mitochondrial DNA used to identify Nicholas. One of these is a female and another is a male. Does that matter? What general statement can you make about their genetic relationship to Nicholas.
9. What was discovered in the mitochondrial DNA of Nicholas that was not identified in either the Duke of Fife or Princess Xenia?
10. What is the term given to the existence of two (or more) genetically different mitochondria in the cell?

Romanov Assignment – C
1. Two “graves” were discovered near Yekaterinburg, Russia. Describe the number of bodies in each grave.
2. When were these graves discovered?
3. One of the reasons that the family of Nicholas II was executed was because there was a fear that the White Russian Army would save them. Who was the White Russian Army?
4. What type of testing was done to confirm sex and familial relationships among the remains found in the mass grave?
5. Genetically, what does STR “stand” for?
6. What three types of DNA were used to test the remains found in a second grave?
7. Of the three types of DNA you listed in #32, which one would have been used specifically to identify Alexis?
8. What was the source of the DNA used to identify Alexis?
9. Was Anastasia in the grave in which Alexis was found?
10. Give a brief history (2-3 sentences) of Anna Anderson-both her claims and what is thought to be true.
11. Where in the US did Anna Anderson eventually settle?
12. Whom did she eventually marry?
13. What were the sources of Anna Andersons’s nuclear DNA?
14. What were the sources of Nicholas’ and Alix’s nuclear DNA?
15. What type of analysis was done on DNA from Anna Anderson, Nicholas, and Alix?
16. Anna Anderson’s mitochondrial DNA was compared to the mitochondrial DNA of what two people?
17. A hypervariable region of the mitochondrial DNA was analyzed. Define a hypervariable region?
18. What were the conclusions from the mitochondrial DNA comparisons?
19. The article which describes the analysis of Anna Anderson’s DNA was published in 1995. When were all of Nicholas’ and Alix’s children finally accounted for?
20. Descibe a current treatment for hemophilia.

Cell Biology
Scientific Literacy – Marine Yeasts
The two yeasts types traditionally studied in cell biology, Saccharomyces cerevisiae (S. cerivisiae) and Schizosaccaromyces pombe (S. pombe), represent the two processes of cell division typically found in yeast organisms. S. cerevisiae divides via budding while S. pombe divides via fission (Saccharomyces cerevisiae 2021; Schizosaccharomyces pombe 2021). Fission and budding are forms of asexual reproduction that are different from mitosis. Furthermore, they are far more different processes than they are alike. One similarity is that they both differ from mitosis. Fission is like mitosis, but they occur in different organisms (Helmenstine 2019). Fission is very rarely seen in eukaryotes while mitosis only occurs in eukaryotes (Helmenstine 2019). Budding on the other hand is mostly seen in eukaryotes (Budding 2021). In fission the parent cell is evenly divided into two indistinguishable daughter cells, while in budding the new cell starts as a small ‘bud’ and grows on the parent cell (Panawala 2017). Budding results in asymmetric reproduction in which the parent cell is larger, while fission, as previously alluded to, results in symmetric reproduction (Panawala 2017). Fission can be observed intracellularly in mitochondria and chloroplasts while intracellular budding occurs in viruses, lysosomes budding from endoplasmic reticulum, and vesicles budding from the plasma membrane (Budding – Budding in Yeast n.d.). Both budding and fission occur in simple organisms, although yeasts, while technically considered simple organisms, are incredibly varied and complex lifeforms.
This discussion covers four marine yeast genera: Candida, Debaryomyces, Cryptococcus, and Rhodotorula; all of which reproduce by budding (Praphailong & Fleet 1999; Hommel 2014; Yeeh 1999; Viviani 2009). Candida is of the Saccharomycetaceae family, is round and has a whitish appearance. Candida normally reproduces via bipolar budding and produces pseudohyphae. It’s found in water, insects, and mammals (Candida (fungus) 2021). Candida is the genus typically responsible for genital yeast infections (Candida (fungus) 2021). Candida species have been used to produce fermented consumables such as wine, sourdough, and sausages, and sometimes causes food spoiling (Hommel 2014). Candida can ferment glucose, maltose, and sucrose, but is unable to ferment lactose (Candida (fungus) 2021). Candida is known to give off a yeasty odor (fungus) 2021). Debaryomyces is also of the Saccharomycetaceae family. It’s typically found in water, soils, foods, and plants, has a round appearance, and can also be infectious to humans (Praphailong & Fleet 1999). Debaryomyces reproduces by multilateral budding (Praphailong & Fleet 1999). Debaryomyces may produce pseudohyphae and produces spheroidal or ovoidal ascospores (Spore produced in a sac called an ascus which is formed by some fungi) (Praphailong & Fleet 1999; Ascus 2021). Sugar fermentation ability varies in strength among Debaryomyces species (Praphailong & Fleet 1999). It is found in meats and cheeses but can spoil fermented foods (Praphailong & Fleet 1999). Debaryomyces has biotechnological potential (biodiesel, surfactants, carotenoids) due to its lipid producing properties (Praphailong & Fleet 1999). Cryptococcus is of the Tramellaceae family and has a glycoprotein outer layer, which gives it its moniker “the sugar yeast” (McDonald, Wiesner & Nielsen 1998). It has an oval to round shape and size can vary from 2-5 micrometers in poorly encapsulated cells and 30-80 micrometers in well encapsulated cells (Viviani 2009). Cryptococcus is usually found in soils, decaying trees, and bird droppings (Viviani 2009). It can be infectious to humans, especially the immunocompromised (Viviani 2009). Cryptococcus does not form hyphae or phseudohyphae (Cryptococcus 2021). Lastly, Rhodotorula is spheroidal, ovoidal, or elongate and of the Sporidiobolaceae family (Rhodotorula 2021; Yeeh 1999). Rhodotorula can form orange, yellow, or reddish colonies (Yeeh 1999). It reproduces by multilateral or polar budding and is non-fermentative (Yeeh 1999). It is among the top ten yeasts that cause food spoilage (Albertyn, Pohl & Viljoen). Rhodotorula is potentially infectious to humans, especially the immunocompromised (Yeeh 1999). Rhodotorula also has bioremediation potential as it has been shown to degrade toxic hydrocarbons in contaminated waters (Rhodotorula 2021). As these general characteristics have shown, yeasts have much variability in appearance, reproduction, habitat, and practical use. The great diversity in yeast species provides an avenue for research and, hopefully, practical solutions to modern problems.
Beginning with qualitative comparisons, three of the four yeast colonies are dark in color: H. werneckii, having a dark grey to amber color, K. petricola, having a mostly dark grey color with a little burgundy, and P. salicorniae, displaying ash to burnt orange and orange colors. A. pullulans differs from the others in that it is not dark at all, but instead a yellow cream color (Mitchison-Field, 2019). Cell shape also varies among H. weneckii, K. petricola, A. pullulans, and P. salicorniae having ovoid, round, lemon, and ovoid/elongate shapes respectively. P. salicorniae colonies however are not well described simply by ovoid and elongate descriptions. They appear as balls of hair formed by roundish cells in the middle with several tentacle-like extensions in all directions (Mitchison-Field, 2019). This exaggerated difference in colony appearance is due to P. salicorniae having both hyphal and yeast cells while the other three only consist of yeast cells. This brings us to growth patterns, which are different in each of the four marine yeasts described by Mitchison-Field et al. Like K. petricola and A. pullulans, H. werneckii divides via budding, however it also alternates budding with fission, unlike the other three yeasts. H. werneckii also prefers alternating over repeating the same division process consecutively (Mitchison-Field, 2019). A. pullulans differs from H. werneckii in that it can form up to six simultaneous buds from the same budding site (Mitchison-Field, 2019). K. petricola also divides via budding but forms linear chains. As mentioned before, P. salicorniae has a bizarre growth pattern. This strange pattern arises from the division septa formed in ovoid and spheroid cells that create triangular compartments and the filamentous hyphae growth. Furthermore, P. salicorniae appears to create a central melanin containing extracellular matrix in which small round cells are produced, creating a sort of meristem (Mitchison-Field, 2019).
Now let’s discuss the more quantitative findings from the information gathered by Mitchison-Field et al. including cell cycle duration, number of nuclei, and time to first bud. Besides having a unique light color, A. pullulans also has various amounts of nuclei in each mother cell and a single nucleus in most daughter cells. The other three yeasts only contain a single nucleus per cell compartment (Mitchison-Field, 2019). The mean cell cycle duration in H. werneckii was 730 minutes and the mean time for a bud to produce a new bud was 253 minutes. It is interesting to note that larger cells had a shorter time until first bud formed. ^Mitchison-Field et al. also noted high plasticity in growth and division of H. werneckii.^ While H. werneckii varies a great deal in time to produce a new bud compared to the traditionally studied yeast S. cerevisiae, K. petricola varied even more in time to produce a bud and cell cycle duration. K. petricola had a mean cell cycle duration of 499 minutes and a time to first bud of 110 minutes and there were outliers that took as little as 10 minutes and as much as 320 minutes until the first bud. Cell cycle duration was also highly varied in K. petricola with lower and upper limits of approximately 250 and 850 minutes. Like H. werneckii and K. petricola, A. pullulans was also highly variable in cell cycle duration but had a significantly lower average cell cycle duration of 159.5 minutes. As mentioned before, A. pullulans sometimes produced up to six buds at once, however these phenomena don’t seem to effect cell cycle duration (Mitchison-Field, 2019). Likely due to the complicated nature and varying cell types found in P. salicorniae, there was no cell cycle duration value given by Mitchison-Field et al. Rather, the authors provided more information regarding cell size at birth, change in cell size, and compared cell area at division for yeast, hyphal, and yeast from hyphae cells.
It’s apparent that the four marine yeasts studied by Mitchison-Field et al. differ from each other a great deal, but they also pose more questions when compared to the yeasts that have been studied in much more detail. The marine yeasts for example seem to have less size control mechanisms when compared to traditional yeast models. Also, unlike S. cerevisiae, which strictly controls singular budding to ensure only one nucleus is distributed to daughter cells, A. pullulans produces several buds at once and has sometimes large and irregularly shaped mother cells. How the nuclei are well distributed many buds remains a mystery. It is also unknown why H. werneckii systematically alternates between budding and fission, but Mitchison-Field et al. infer it creates a situation in which subsets of the population can deal with sudden stressors that may occur in the marine environment. Lastly, the curious triangular cell compartments that arise in P. salicorniae are much unlike traditional fungal models (Mitchison-Field, 2019). Overall, the marine yeast study by Mitchison-Field et al. compiled an array of data that can be used and compared with the future research of yeasts no doubt inspired by this study and verified the ever complicated and variable nature of yeasts. Much is yet to be understood.
| H. werneckii | K. petricola | A. pullulans | P. salicorniae | |
| Mean cell cycle duration | 730 min | 499 min | 159.5 min | n/a |
| Mean time to first bud | 253 min | 110 min | n/a | n/a |
| Growth pattern | Alternating fission/budding | Budding -forms chains | Multiple simultaneous budding | Central cluster of cells with division septa, sometimes results in triangular cell compartments (meristematic) and hyphal growth, produces fuzzy looking central cluster with tentacle-like filamentous extensions |
| Hyphal or yeast cells | yeast | yeast | yeast | both |
| # nuclei | Single nucleus per cell compartment | Single nucleus per cell | Variable # of nuclei in mother cell, buds typically with single nucleus | Single nucleus per cell compartment |
| Cell shape | Ovoid | spherical | Apiculate or lemon shape | Ovoid/elongate |
| Colony color | Dark gray to amber | Dark gray with little burgundy | Yellow/cream color | Ash to burnt orange/orange color |
References
Mitchison-Field et al., 2019, ‘Unconventional Cell Division Cycles from Marine-Derived Yeasts’, Current Biology, vol. 29, pp. 3439-3456, doi:10.1016/j.cub.2019.08.050
Saccharomyces cerevisiae 2021, Wikipedia, viewed 14 November 2021, https://en.wikipedia.org/wiki/Saccharomyces_cerevisiae
Schizosaccharomyces pombe 2021, Wikipedia, viewed 14 November 2021, https://en.wikipedia.org/wiki/Schizosaccharomyces_pombe
Helmenstine, A 2019, Binary Fission vs. Mitosis, viewed 14 November 2021, https://www.thoughtco.com/binary-fission-vs-mitosis-similarities-and-differences-4170307
Budding 2021, Wikipedia, viewed 14 November 2021, https://en.wikipedia.org/wiki/Budding
Panawala, L 2017, Difference Between Binary Fission and Budding, viewed 14 November 2021, https://pediaa.com/difference-between-binary-fission-and-budding/
Budding – Budding in Yeast and Budding in Hydra n.d., Vedantu, viewed 14 November 2021, https://www.vedantu.com/biology/budding
Praphailong, W & Fleet, G 1999, ‘Debaryomyces’, in R Robinson (ed.), Encyclopedia of Food Microbiology, Elsevier, pp. 515-520
Hommel, RK 2014, ‘CANDIDA | Introduction’, in C Batt & ML Tortorello (eds), Encyclopedia of Food Microbiology, 2nd edn, Academic Press, pp. 367-373
Yeeh, Y 1999, ‘Rhodotorula’, in R Robinson (ed.), Encyclopedia of Food Microbiology, Elsevier, pp. 1900-1905
Viviani, M, Tortorano, AM 2009, Clinical Mycology, 2nd edition, Churchill Livingstone, viewed 14 November 2021, https://www.sciencedirect.com/science/article/pii/B9781416056805000098
Candida (fungus) 2021, Wikipedia, viewed 14 November 2021, https://en.wikipedia.org/wiki/Candida_(fungus)
Ascus 2021, Wikipedia, viewed 14 November 2021, https://en.wikipedia.org/wiki/Ascus
McDonald, T, Wiesner, DL & Nielsen, K 1998, ‘Cryptococcus’, Current Biology, vol. 22, no. 14, pp. 554-555
Cryptococcus 2021, Wikipedia, viewed 14 November 2021, https://en.wikipedia.org/wiki/Cryptococcus
Rhodotorula 2021, Wikipedia, viewed 14 November 2021, https://en.wikipedia.org/wiki/Rhodotorula
Albertyn, J, Pohl, CH & Voljoen, BC 2009, Clinical Mycology, 2nd edition, Churchill Livingstone, viewed 14 November 2021, https://www.sciencedirect.com/science/article/pii/B9780123847300002895

Cell Biology end of term reflection:
Learning about insulin binding to RTK, and the subsequent intracellular events, was fascinating and gave me a deeper understanding of insulin function alongside what I learned in Anatomy & Physiology 2 this semester. Although my understanding isn’t complete, I now understand the basic concept that insulin binding activates protein phosphatases, thereby stimulating the production of glycogen by glycogen synthase. As an aspiring physician assistant, I’ve often wondered about signaling pathways/cascades without knowing the terminology to even ask the question. Although I had some difficulty with the work load this semester, I learned a great deal in Cell Biology, and I’m now more prepared to expand on what I’ve learned and admittedly to refresh what may have slipped my mind.