Glimmer to analyze CDSs in a partial sequence
For this example, I will use the Spiroplasma helicoides strain TABS-2, whose genome was submitted to GenBank on August 23, 2016.
The first portion will require a training file. This will be the whole genome of Spiroplasma helicoides strain TABS-2.
This will enable the program to understand the genome and the way to deal with my data input and the required output.
My second file contains the partial sequence with CDS locations .
Training and determining determine CDS locations
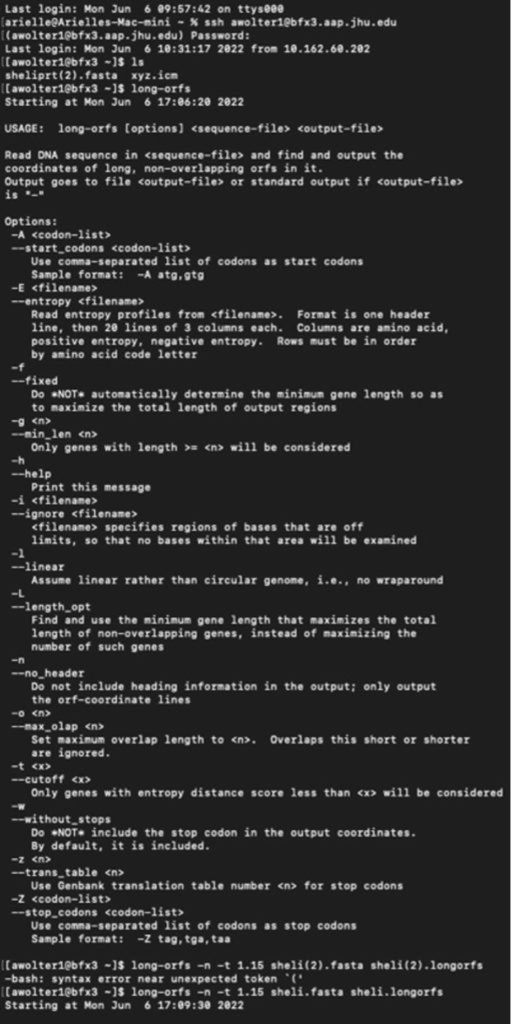
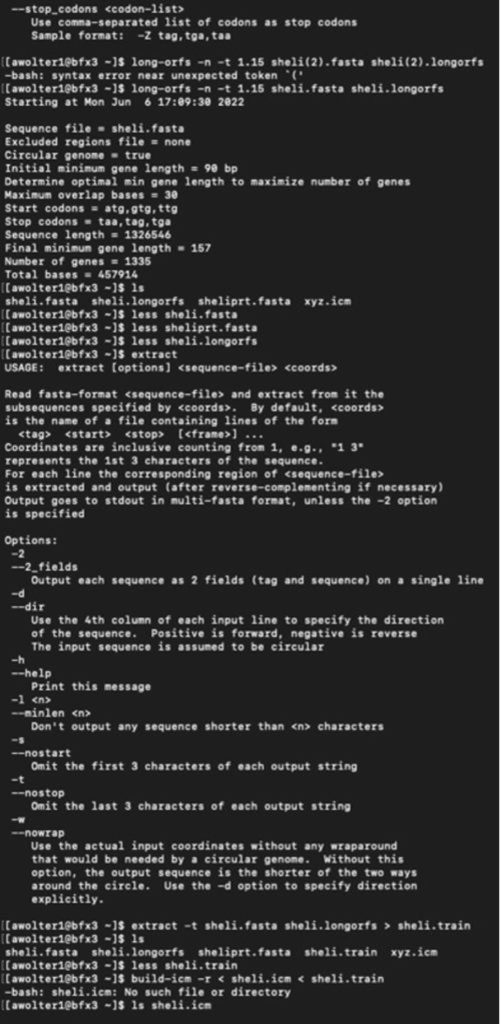
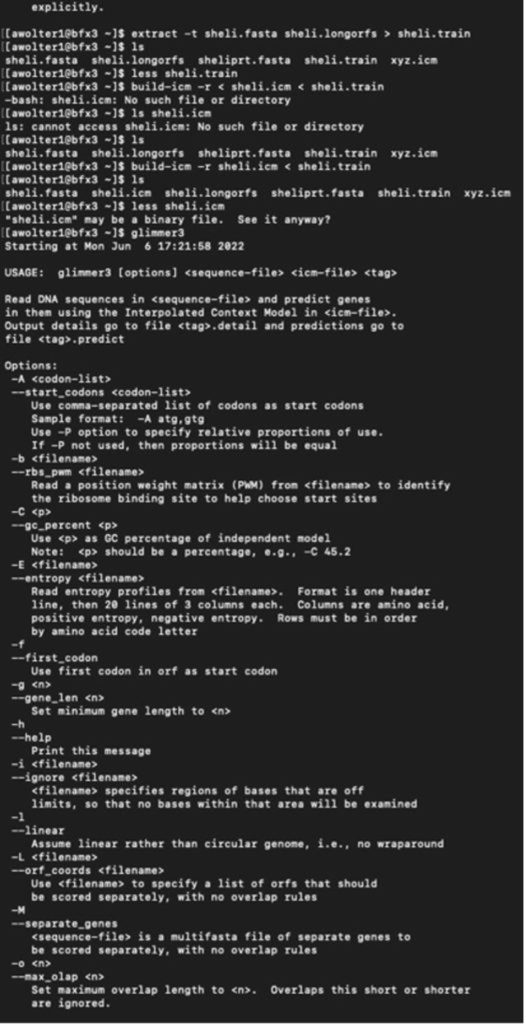
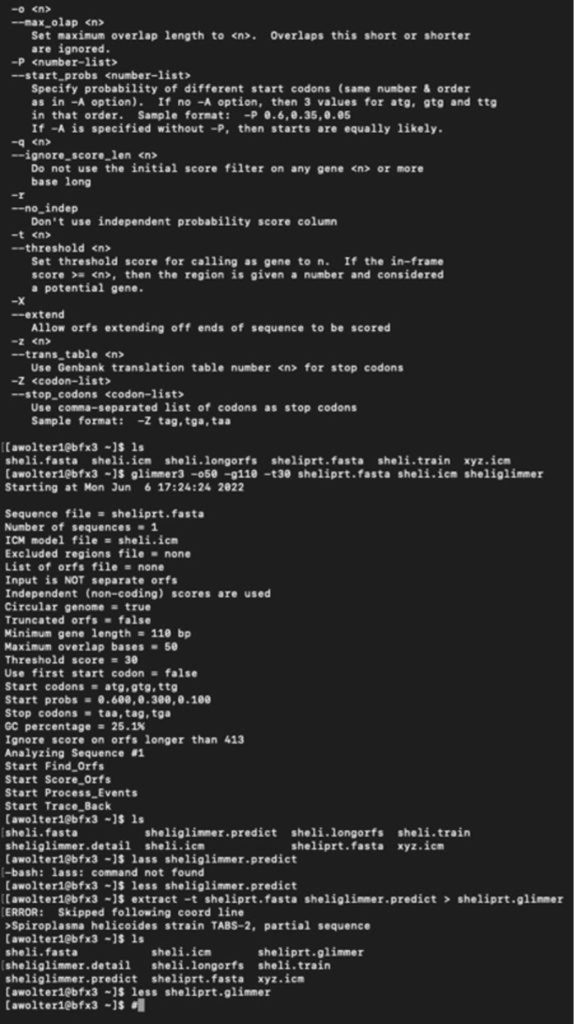
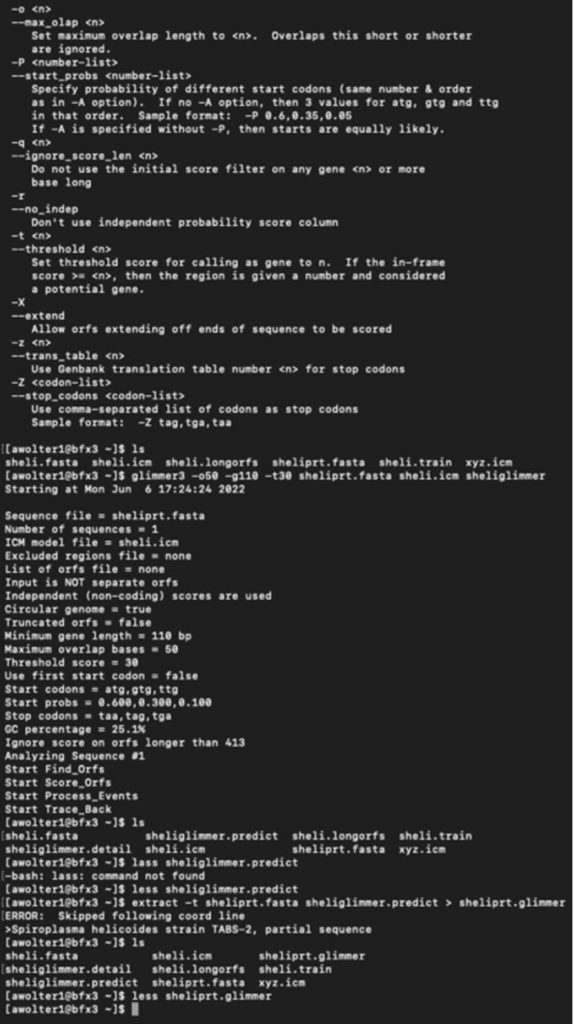
I have logged into the BFX server and uploaded the full and partial files so I can work with GLIMMER in terminal:
Below you can find a partial list of commands to retrieve the orbs below. For more detailed information please see the work flow from terminal below:
Training Set:
full genome=> sheli.fasta Prediction sequence: partial genome => sheliprt.fasta [awolter1@bfx3 ~]$ less sheliglimmer.predict [awolter1@bfx3 ~]$ extract -t sheliprt.fasta sheliglimmer.predict > sheliprt.glimmer >Spiroplasma helicoides strain TABS-2, partial sequence [awolter1@bfx3 ~]$ ls sheli.fasta sheli.icm sheliprt.glimmer sheliglimmer.detail sheli.longorfs sheli.train sheliglimmer.predict sheliprt.fasta xyz.icm [awolter1@bfx3 ~]$ less sheliprt.glimmer [awolter1@bfx3 ~]$ less sheliglimmer.predict >Spiroplasma helicoides strain TABS-2, partial sequence orf00001 635 991 +2 4.13 orf00002 998 1141 +2 4.42 orf00003 1154 1312 +2 2.30 orf00004 1334 1978 +2 5.68 orf00006 2242 2463 +1 6.25 orf00008 2585 4003 +2 8.80 orf00009 4010 4678 +2 8.48 orf00010 4880 5143 +2 6.98 GLIMMER Session in Terminal: