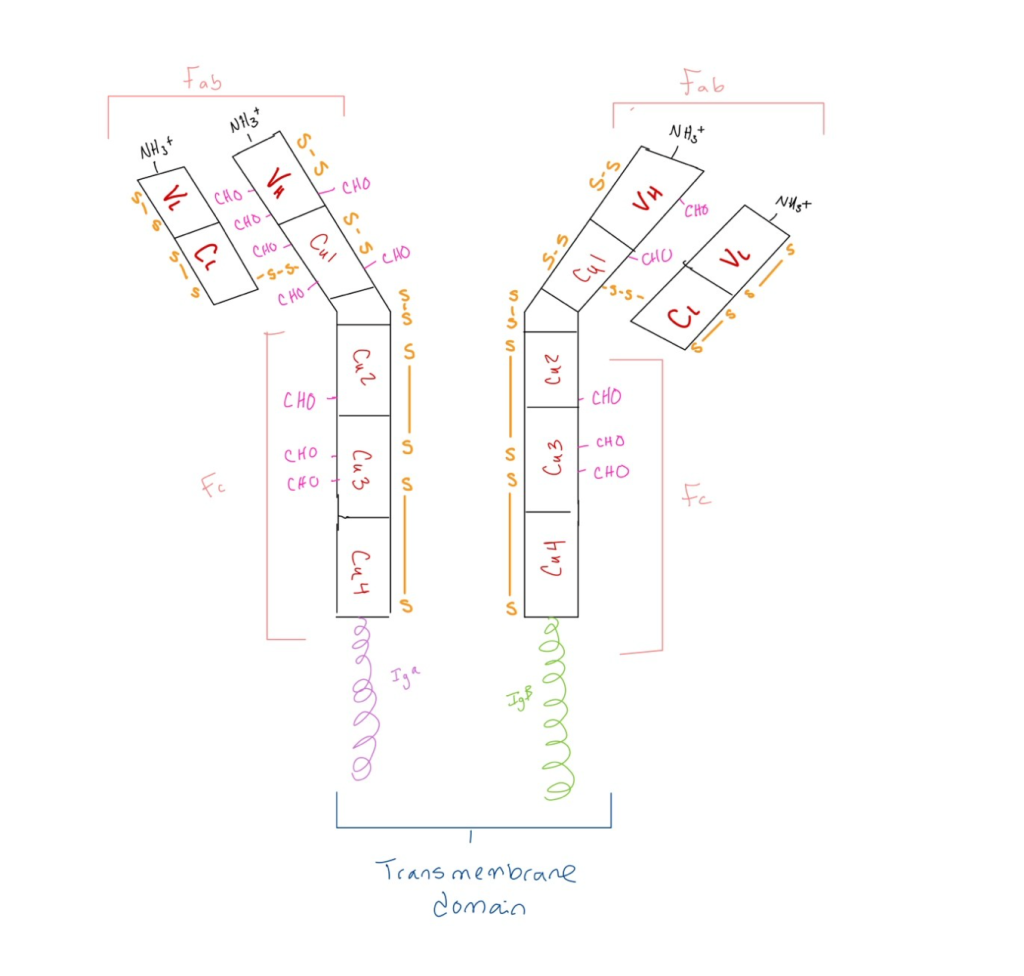
One important concept I learned in my immunology course that helped me make a connection to my specific interest in cytology was the role of cell signaling in immune responses. In immunology, I studied how immune cells, such as T-cells and B-cells, communicate to coordinate the body’s defense against pathogens. Understanding how immune cells rely on signaling molecules to activate or suppress immune responses helped me see the parallels between immune system regulation and cellular homeostasis. It also highlighted the complexity of cellular communication, which is crucial for both immune function and normal cellular behavior. Learning these concepts allowed me to better understand how dysfunctions in cell signaling can lead to diseases, such as autoimmune disorders or cancer. This connection deepened my understanding of immunology and strengthened my ability to approach biological problems from a more integrated perspective.
Author: emaye002
-mab Drug Assignment
Daratumumab is a monoclonal antibody that treats myeloma by targeting the CD38 protein on myeloma cells (International Myeloma Foundation, 2024). Myeloma is a form of cancer of the plasma cells (American Society of Hematology, 2024). Plasma cells are a type of white blood cell that help the body fight infections. Myeloma cells prevent the normal production of antibodies, overall weakening the immune system. Myeloma cells also release substances that break down the bones (American Society of Hematology, 2024). Myeloma cells originate in the bone marrow and travel through the bloodstream, accumulating in bones throughout the body. Common sites of accumulation include the spine, skull, ribcage, and pelvis. Myeloma in the bone marrow causes a number of complications such as anemia (low red blood cell count), neutropenia (low white blood cell count), and thrombocytopenia (low platelet count) (International Myeloma Foundation, 2024). Kidney disease is complication that comes about in around 50% of patients with active myeloma. This is because of the toxicity of the monoclonal proteins produces by myeloma cells (International Myeloma Foundation, 2024). Since these myeloma cells affect multiple areas of bone marrow, the disease is often called multiple myeloma. In multiple myeloma, the plasma cells are effectively transformed into malignant cancerous cells which then produce an abnormal antibody called M protein (Multiple Myeloma Research Foundation, 2024). An abundance of M protein is a defining characteristic of myeloma. Multiple myeloma is triggered by specific genetic mutations which can be different from person to person. However, myeloma is not considered to be hereditary, these mutations can occur randomly as people grow older (Multiple Myeloma Research Foundation, 2024).
Daratumumab is an IgG antibody.
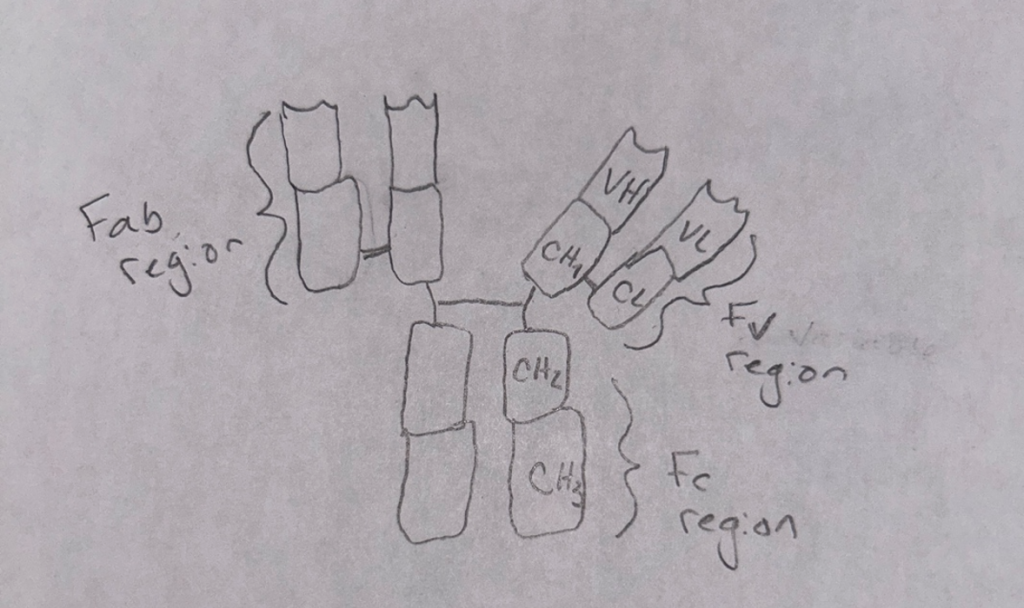
Researchers found that daratumumab has an effect on the immune system. Studies showed that daratumumab helps boost the immune response by eliminating certain immune cells that suppress the body’s ability to fight tumors (Plesner & Krejcik, 2018). These cells, which express a protein called CD38, belong to the T cell, B cell, and monocyte–macrophage systems. By removing these immunosuppressive cells, daratumumab allows cytotoxic T cells to grow and work better, which is linked to better survival rates in patients, even if their M-protein levels don’t drop significantly. Additionally, daratumumab seems to improve the activity of these T cells by blocking the CD38 protein’s ability to produce immunosuppressive molecules like adenosine, which weakens the immune response (Plesner & Krejcik, 2018). This boost in T cell function helps control the disease more effectively. CD38 can be found on myeloma cells, cells around them, or even in tiny vesicles released by myeloma cells, which can contribute to treatment failure by reducing the drug’s effectiveness. It’s also been suggested that myeloma cells with low levels of CD38 after starting daratumumab treatment might not respond well (Plesner & Krejcik, 2018). To address this, researchers are testing ways to increase CD38 levels on these cells. In a model of non-small cell lung cancer, research has shown that CD38 helps cancer cells grow and survive. This may be the case for myeloma, where high levels of CD38 could help myeloma cells live longer (Plesner & Krejcik, 2018). On the other hand, when treatment with daratumumab lowers CD38 levels, it might make the myeloma cells more sensitive to other anti-myeloma treatments.
References
Myeloma cancer questions FAQ | International Myeloma Foundation. International Myeloma Foundation. (2024). https://www.myeloma.org/myeloma-cancer-questions
Myeloma. American Society of Hematology. (2024). https://www.hematology.org/education/patients/blood-cancers/myeloma#:~:text=Myeloma%20is%20cancer%20of%20the,weakened%20and%20susceptible%20to%20infection.
Plesner, T., & Krejcik, J. (2018). Daratumumab for the Treatment of Multiple Myeloma. Frontiers in immunology, 9, 1228. https://doi.org/10.3389/fimmu.2018.01228
What is multiple myeloma? symptoms, causes, & prognosis. Multiple Myeloma Research Foundation. (2024, May 29). https://themmrf.org/multiple-myeloma/
Favorite Food Writeup
Fats
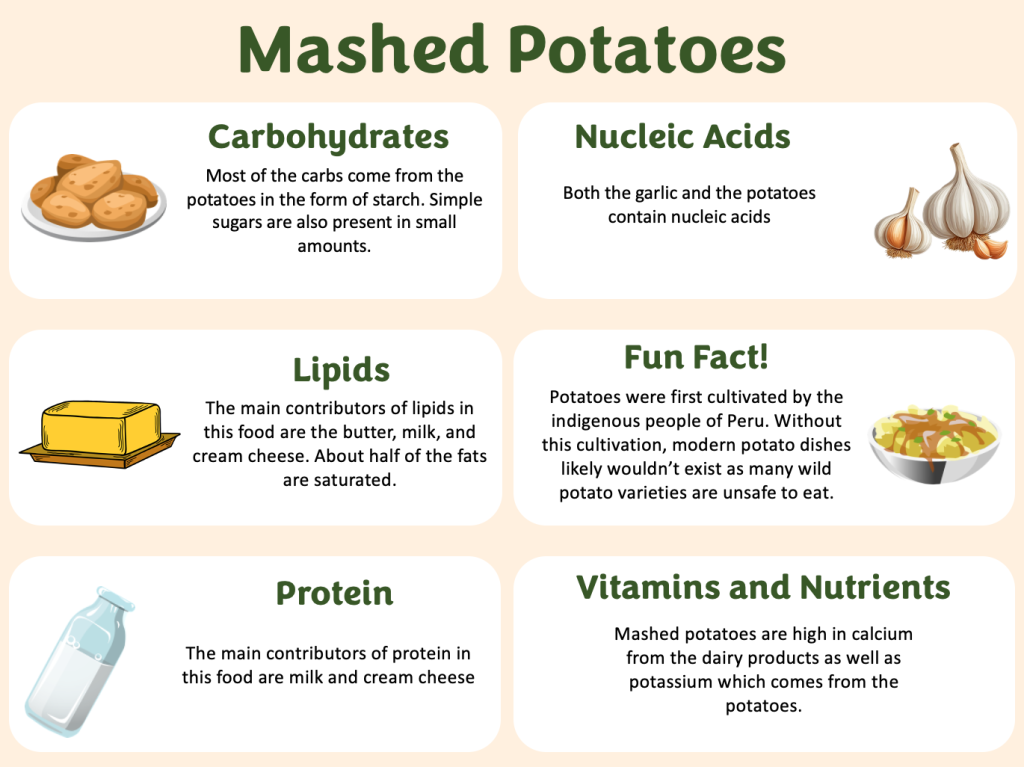
The three main contributors of fats in mashed potatoes are milk, cream cheese, and butter. Milk contains myristic, stearic, and palmitic acids. Butter and cream cheese contain mostly oleic, myristic, and palmitic acids.
- Myristic acid – 14 carbons, saturated, nonessential
- Stearic acid – 18 carbons, saturated, nonessential
- Palmitic acid – 16 carbons, saturated, nonessential
- Oleic acid – 18 carbons, mono-unsaturated, omega-9, nonessential
The most common fatty acid between all of these foods is palmitic acid, pictured below:
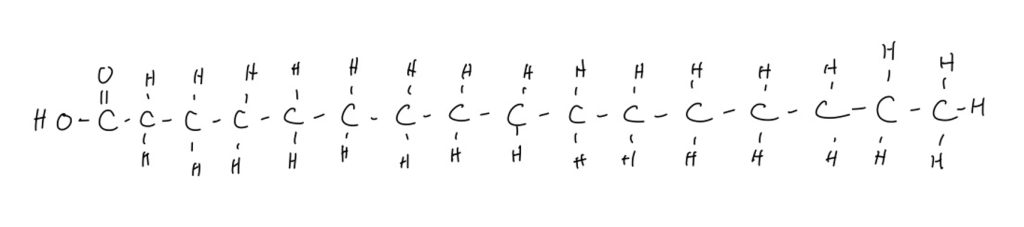
Carbohydrates
The main source of carbohydrates in mashed potatoes is the starch in the potatoes. There is also carbohydrates in milk in the form of lactose. These are both natural sugars.
Protein
The main source of protein in mashed potatoes is milk. There are 8 grams of protein per cup in whole milk. Milk is a good source of the amino acid leucine.
Vitamins and Minerals
Vitamin D (from milk) – Good source, 10%
Calcium (from milk) – Excellent source, 20%
Vitamin A (from milk) – Good source, 10%
Potassium (from potatoes) – Excellent source, 20%
Iron (from potatoes) – Good source, 10%
DNA Privacy Policy Writeup
Ancestry DNA and 23 and Me are both companies that offer DNA testing kits for people to have genetic testing done. However, both companies have had public scandals revolving around selling user data and data leaks. So, how do they protect user data? 23 and Me states that they conduct all analyses with de-identified data, so the genomic information and personal identifying information are separate. They also do not publish any research information that identifies any particular individuals. Ancestry DNA claims to use secure software to encrypt all user data including personal and genetic information. However, they don’t mention any specifics about de-identifying the data stored at all. It can be implied that PII and genetic information are linked in their records. 23 and me outwardly states that all data including personal information can be sold in the case of mergers or bankruptcy. Ancestry also states that the information in their databases can be sold under those circumstances as well as sold to analytics and advertising companies for targeted ads. They state that de-identified data is shared with research partners with user consent.
With the rise of these genomic databases there have been plenty of news stories about law enforcement using them to match forensic DNA to website users. There has also been some concern that these websites will share data with health insurance companies, causing higher rates for those with an increased likelihood of illnesses. Both websites have the same stance on sharing information in these cases. Law enforcement needs a search warrant, a subpoena, or a court order to access their databases. As for health insurance companies, information will not be shared unless there is expressed consent from the user. When it comes to data retention and deletion, things get kind of murky in the privacy policies. 23 and Me gives users the option to either have their sample stored or destroyed after analysis. Users can also delete their account however there is no guarantee of data removal after you do so. They state that they have a legal requirement to retain all information for an unspecified period of time before it can be removed. Ancestry DNA has a more cut and dry answer, DNA samples are kept for seven years after which they can be destroyed, and genetic/personal information will be removed from their database. However, ‘usage’ information is retained seemingly indefinitely even after account deletion. If one chooses to opt into having their information used for additional research, it is used about the same for each company. Genetic information can be used for research into diseases, traits, and population history both by the company itself and third-party collaborators.
Personally, I would be hesitant to fully entrust my genetic information to either company. With that being said, I would be more inclined to use 23 and Me. Both companies allow the sharing or sale of data under specific circumstances. Even if data is de-identified, there is a risk that advancements in technology could allow for re-identification. The fact that both companies retain data even after account deletion, with Ancestry DNA keeping usage data and 23 and Me holding data for an undefined “limited period,” limits control over your personal information once shared. 23 and Me has better de-identification measures when it comes to research, which suggests a stronger emphasis on privacy for research purposes. Ancestry DNA has a lack of clear separation between PII and genetic data, which is a potential vulnerability. They also explicitly mention that they share data with advertising and analytics companies. Neither company offers complete protection but 23 and Me seems to have better policies when it comes to protecting your data. However, I personally wouldn’t use either website due to the inherent risks of putting personal information out there.
Genome Assignment
Chromosome Maps
1. WHAT CHROMOSOME DID YOU CHOOSE?
Chromosome 12
2 & 3. STATE THE NUMBER OF GENES AND BASE PAIRS ON THE CHROMOSOME YOU CHOSE.
1600 genes and 130 million base pairs
4. LIST ONE GENE THAT IS LOCATED ON THIS CHROMOSOME.
PAH gene.
5. STATE THE FUNCTION OF THE GENE YOU LISTED IN #42
The gene for phenylalanine hydroxylase (PAH) converts phenylalanine to tyrosine.
Introduction to Nucleotide BLAST
6. WHAT IS THE SECOND SEQUENCE DESCRIPTION MATCH FOR YOUR QUERY SEQUENCE?
Homo sapiens CFTR (CFTR) gene, partial cds
7. WHAT DOES THE ENCODED PROTEIN DO IN THE BODY?
It functions as a chloride channel, controlling ions and water secretion and absorption in epithelial tissues.
8. FOR WHAT DISEASE IS A MUTATED FORM OF THIS GENE RESPONSIBLE?
Cystic fibrosis
9. ON WHAT CHROMOSOME IS THE GENE LOCATED?
Chromosome 7
10. Return to the original nucleotide sequence alignment descriptions. CHOOSE A SPECIES (STATE THE SCIENTIFIC NAME) OTHER THAN HOMO SAPIENS THAT ALSO HAS A 100% IDENTITY (Per. Ident) FOR THIS SEQUENCE?
Pongo abelii
11. WHAT IS THE COMMON NAME FOR THIS SPECIES?
Sumatran orangutan
12. DOES IT SURPRISE YOU THAT THIS SPECIES ALSO HAS A 100% SIMILARITY IN IDENTITY? WHY OR WHY NOT?
It does not as orangutans are also primates and share common ancestry with homo sapiens.
13.
a. WHAT IS THE GENUS AND SPECIES WITH THIS NUCLEOTIDE SEQUENCE?
Sapajus apella
b. WHAT IS THE COMMON NAME?
Tufted capuchin
c. ARE THERE ANY GAPS BETWEEN THE TWO SEQUENCES (THE ONE YOU ORIGINALLY SUBMITTED AND ONE THAT HAS LESS THAN 100% QUERY COVER)?
Yes, the original has 0/120 and the other has 1/119.
14. WHAT IS A GAP IN SEQUENCE ALIGNMENTS?
Gaps in sequence alignments are used to account for mutations that occur from insertions or deletion in the sequence.
FOR EACH, STATE WHAT THE GENE IS (#15-18). (Again, give the description of the gene or gene product, not the nucleotide sequence.)
15. NM_145556
Mus musculus TAR DNA binding protein
16. NM_013444
Homo sapiens ubiquilin 2
17. NM_001010850
Homo sapiens FUS RNA binding protein
18. KJ174530
Homo sapiens superoxide dismutase-1 (SOD-1) gene
19. WHAT DISEASE IS ASSOCIATED WITH MUTATIONS OF THE GENES REFERENCED IN #15-#18? WHAT IS A “COMMON NAME” OF THE DISEASE?
Amyotrophic lateral sclerosis (ALS), commonly called Lou Gehrig’s disease
20. WHAT IS GENBANK?
GenBank is a database from the NIH which is a collection of all DNA sequences that are publicly available.
Introduction to Protein BLAST
21. First, answer this question: WHAT IS cDNA?\
cDNA is complementary DNA that was reverse transcribed from RNA.
22. WHAT IS THE SEQUENCE MATCH?
Beta-globin
23. DO YOU SEE ANY DIFFERENCES BETWEEN THE TWO AMINO ACID SEQUENCES?
Yes
24. IF YOU SAW DIFFERENCES, WHAT WERE THEY?
There is one gap where the query sequence is missing an ‘R’ where the subject has one. There is another where the query is missing an ‘S’ where the subject has one
25. ARE THERE ANY GAPS IN THE SEQUENCE ALIGNMENT?
Yes, 1/806
26. WHAT GENE ENCODES FOR THE POLYPEPTIDE YOU WERE ANALYZING?
Fibroblast growth factor receptor 3
27. WHAT IS THE FUNCTION OF THIS PROTEIN?
Interacts with fibroblast growth factors and influences mitogenesis and differentiation. It binds with the fibroblast growth hormone and plays a role in bone development and maintenance.
28. WHAT HUMAN DISEASE IS CAUSED BY A MUTATION IN THIS GENE?
Craniosynostosis and skeletal dysplasia.
29. WHAT IS THE CONNECTION AMONG THE FOLLOWING: NIH, NLM, NCBI, and HHS?
The National Library of Medicine (NLM) and The National Center for Biotechnology Information (NCBI) are both a part of and overseen by the National Institutes of Health (NIH). The NIH falls under the U.S Department of Health and Human Services (HHS) and is overseen by them.
30. WHAT WAS ONE POSITIVE THING AND ONE NEGATIVE THING YOU ENCOUNTERED WHILE DOING THIS ASSIGNMENT?
One positive I encountered while doing this assignment was getting to explore the genomic sequencing databases and learn a lot about all of the information available on the websites provided. One negative thing I encountered was navigating the information that went over my head in order to actually find what I was looking for.
Lac Operon Assignment
1. When lactose is absent, the lacl produces a repressor which binds to the DNA of the operator, preventing gene expression.
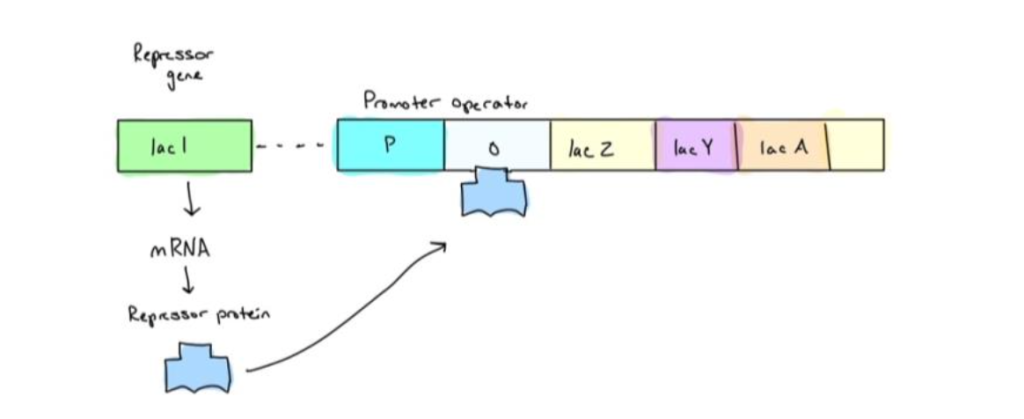
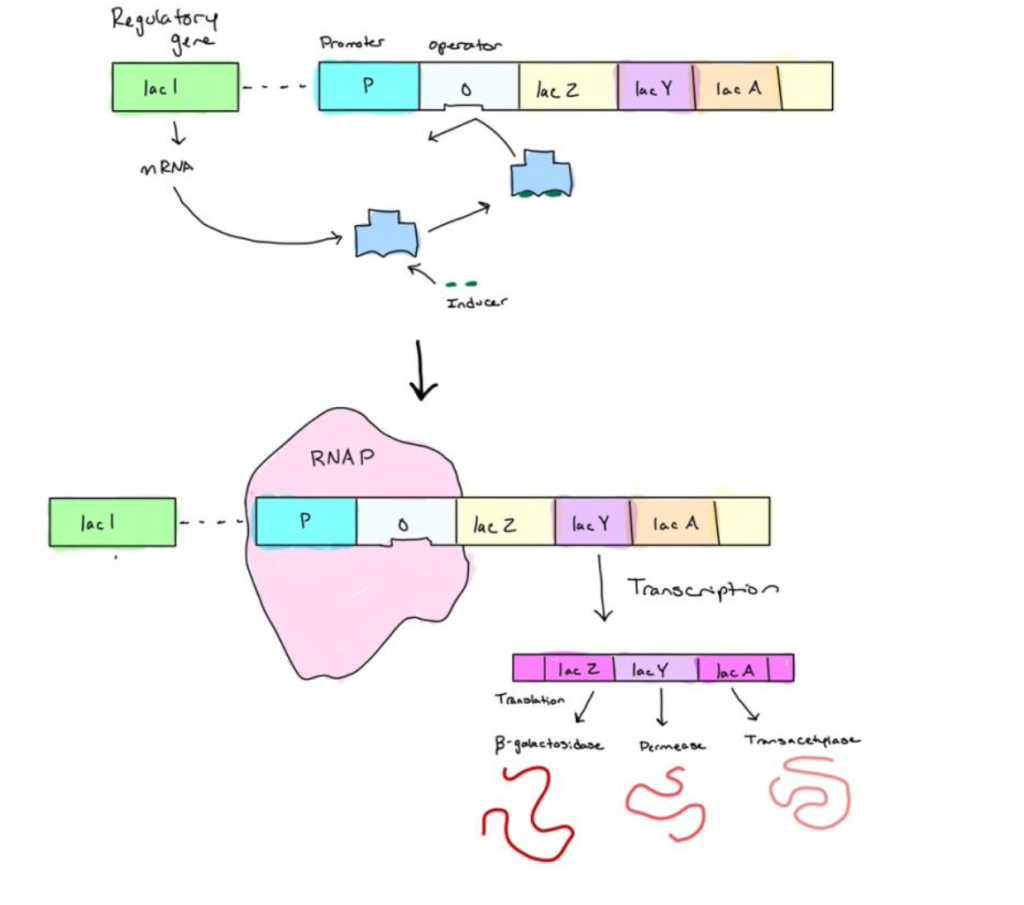
2. When lactose is present, it acts as an inducer and binds to the repressor. Therefore, the repressor cannot bind to the operator. RNA polymerase now has access to the promoter and initiates the transcription of the three lactose-utilization genes.
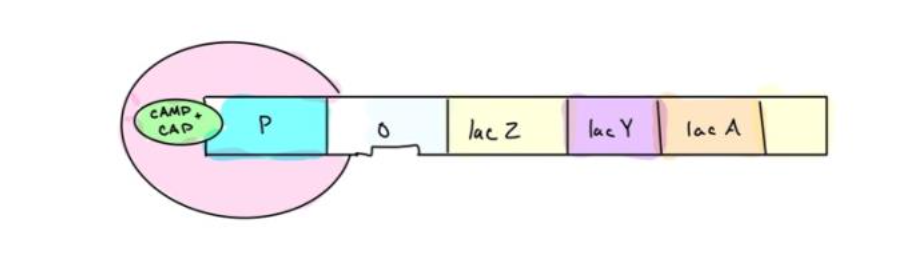
3. When glucose is absent, CAP (activator) is active due to high levels of cAMP is present.
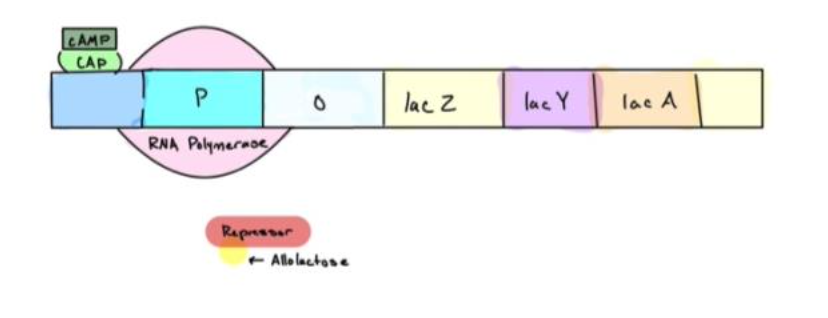
4. When glucose is absent, but lactose is present, cAMP levels are high therefore CAP is active and bound to the DNA of the promoter. CAP allows RNA Polymerase to bind to the promoter. The lac repressor is inactive and strong transcription can occur.
5. The lac operon is regulated during the transcriptional stage of gene expression.
Written Assignment #5 – Genetics Related Article from the Popular Press
In an NBC News article titled” A quick test could protect against fatal chemo overdose, yet few doctors use it,” Arthur Allen tells the story of a 70-year-old woman who tragically died after an overdose of a chemotherapy drug. The drug she was prescribed is a pill called capecitabine which is chemically similar to the intravenous chemotherapy drug known as 5-FU or fluorouracil. While these drugs are used commonly in cancer treatment, those who are deficient in an enzyme that metabolizes the drugs can experience a slew of unpleasant symptoms or, in Carol Rosen’s case, die.
In his article, Allen states that the reason patients die from chemotherapy drugs is because the drugs stay in the body for hours instead of being quickly metabolized. In a review titled” Testing for Dihydropyrimidine Dehydrogenase Deficiency to Individualize 5-Fluorouracil Therapy,” the authors go into more detail about the way that the molecular mechanisms work for an overdose to happen. Genetic factors are a big contributor to the risk of developing severe toxicity to 5-FU. This was confirmed by researchers through advanced pedigree analyses (Diasio & Offer, 2022). The enzyme necessary for metabolizing the drugs is dihydropyrimidine dehydrogenase (DPD) and two deleterious variants were identified in the gene that encodes DPD. The genes segregate independently and have an autosomal codominant inheritance pattern (Diasio & Offer, 2022). Since then, many other variants have been identified after more clinical testing. In individuals with DPD deficiency, it causes the catabolic pathway to shift toward anabolism, potentially causing severe toxicity. However, not all of those with the deficiency will be at severe risk if they use the topical forms of the chemotherapy drugs. Arthur Allen did a good job of simplifying the way that the overdose is caused, but Diasio and Offer went more into depth about what happens at a cellular level.
Due to the correlation between DPD deficiency and 5-FU toxicity, it is important to give doctors ways to identify DPD deficiency so that adjustments to the dosage of the medication can be made. There are both phenotypic tests and genotype-based tests that can be performed to test for DPD deficiency. Genotypic tests have higher accuracy and have been used frequently in European countries (Diasio & Offer, 2022). After her mother’s death, Carol Rosen’s daughter urged the hospital to implement genetic testing before prescribing fluoropyrimidine. The hospital quickly adopted a system to test patients and around 50 patients were detected with the deficiency within 10 months (Allen, 2024). Multiple different tests can be used; some check for eleven potentially dangerous variants, whereas others only screen for eight or four. An important factor in these genetic tests is that they are not one size fits all and different tests may be necessary for different ancestries. Most of the clinical studies have been conducted in Europe but different variants of the enzyme deficiency have been found in African American individuals that did not appear in European patients (Diasio & Offer, 2022). A man named Dr. Anil Kapoor died as a result of fluoropyrimidine poisoning. Kapoor was tested for four variants of the deleterious gene, but he carried a variant of the gene associated with South Asian descent that is not covered in the European-based genetic tests (Allen, 2024). While tests that screen for hundreds of variants are out there, they can be expensive, and getting results back can take longer. Some doctors have pushed back on mandatory testing, stating that the results can be unclear and lead to undertreatment of patients. Fluorouracil is not the only drug that has been prescribed to people who lack the enzyme to properly metabolize it. An anti-blood clotting medication called clopidogrel was marketed as safe for Native Hawaiians while more than 50% of them lack an important enzyme that processes clopidogrel.
References
Allen, A. (2022, March 27). A quick test could protect against fatal chemo overdose, yet few doctors use it. NBCNews.com. https://www.nbcnews.com/health/cancer/quick-test-protect-fatal-chemo-overdose-yet-cancer-doctors-use-rcna144664
Diasio, B. Offer, S. Testing for Dihydropyrimidine Dehydrogenase Deficiency to Individualize 5-Fluorouracil Therapy. MDPI; https://www.mdpi.com/2072-6694/14/13/3207 (2022).
Written Assignment 4: Primary Genetics Article Review
In the article titled,” Transcriptome-directed analysis for Mendelian disease diagnosis overcomes limitations of conventional genomic testing,” researchers aim to improve the diagnosis of rare genetic diseases by using transcriptome sequencing. By focusing on the patterns of gene activity, the researchers were able to diagnose previously unsolved cases. This type of sequencing can improve diagnostic rates by 7.5% to 36% depending on the tissue sampled and the patient’s condition. Rather than looking for specific genetic changes upfront, the methods used here searched for unusual patterns in gene activity that could point to underlying genetic issues. As a result, they were able to diagnose 12% of their cases overall, and 17% when excluding cases that were already answered by standard genetic testing. This approach looks specifically at how genes are being used to understand what may be going wrong at the genetic level. This method is distinct from typical genetic testing, which begins with looking at individual genetic abnormalities and attempts to match them with observed traits or phenotypes. Instead by focusing on gene activity patters, transcriptome sequencing can uncover issues that may be missed by standard testing. Such as noncoding genetic agents or small variations in DNA.
In this study, the researchers used RNA sequencing as the first step in their process. They analyzed the gene activity patterns in patients’ blood and skin cells to guide their genetic analysis. While other studies have used this approach, this is the first study to apply this method to a wide range of conditions in patients. By analyzing the gene activity in both whole blood and fibroblast cells, which are commonly used in clinical research, they found that fibroblast cells in particular were more informative for a variety of conditions, especially those involving the nervous system. This is because they showed more consistent gene expression than whole blood, making them better for detecting relevant differences in gene expression. The researchers were also able to highlight the limitations of standard genetic testing such as exome sequencing and chromosomal microarray analysis. These tests have the potential to miss important genetic changes, particularly in the noncoding regions of DNA or when the changes are very small. The transcriptome-directed approach, which combines RNA sequencing with genetic analysis, proved to be more effective in finding these missed variations.
Several cases proved to be very important. Including one in which RNA sequencing identified a genetic variant missed by other testing, leading to the diagnoses of Renpenning syndrome. In another case, RNA sequencing showed a deletion in CLTC gene, causing intellectual disability. In other cases, KANSL1 and NSD2 gene abnormalities were detected through RNA sequencing, leading to the diagnosis of Koolen-de Vries syndrome and NSD2-associated intellectual disability syndrome, respectively.
In conclusion, this study shows thar analyzing gene activity patterns is effective in diagnosis rare Mendelian diseases that standard genetic testing may overlook. By focusing on how genes are being used, specifically in fibroblast cells, researchers can uncover genetic issues that traditional tests overlook. This approach could significantly improve the diagnostic rates for patients with challenging conditions. What was unique about this study is the diverse group of patients with various symptoms and age ranges. The researchers believe that integrating RNA sequencing into standard genetic testing will help diagnoses more patients, especially those who cannot be diagnoses through exome sequencing or chromosomal microarray analysis.
References
Murdock, D. et al. Transcriptome-directed analysis for Mendelian disease diagnosis overcomes limitations of conventional genomic testing. The Journal of Clinical Investigation; https://doi.org/10.1172/JCI141500 (2021).