Writing Assignment 1
Writing Assignment 2
Shen, Z., Xiao, Y., Kang, L., Ma, W., Shi, L., Zhang, L., Zhou, Z., Yang, J., Zhong, J., Yang, D., Guo, L., Zhang, G., Li, H., Xu, Y., Chen, M., Gao, Z., Wang, J., Ren, L., & Li, M. (2020). Genomic diversity of severe acute respiratory syndrome-coronavirus 2 in patient with coronavirus disease 2019. Oxford Academic, 71(15), 713-720. doi: https://doi.org/10.1093/cid/ciaa203
Writing Assignment 3
Writing Assignment 4
Writing Assignment 5
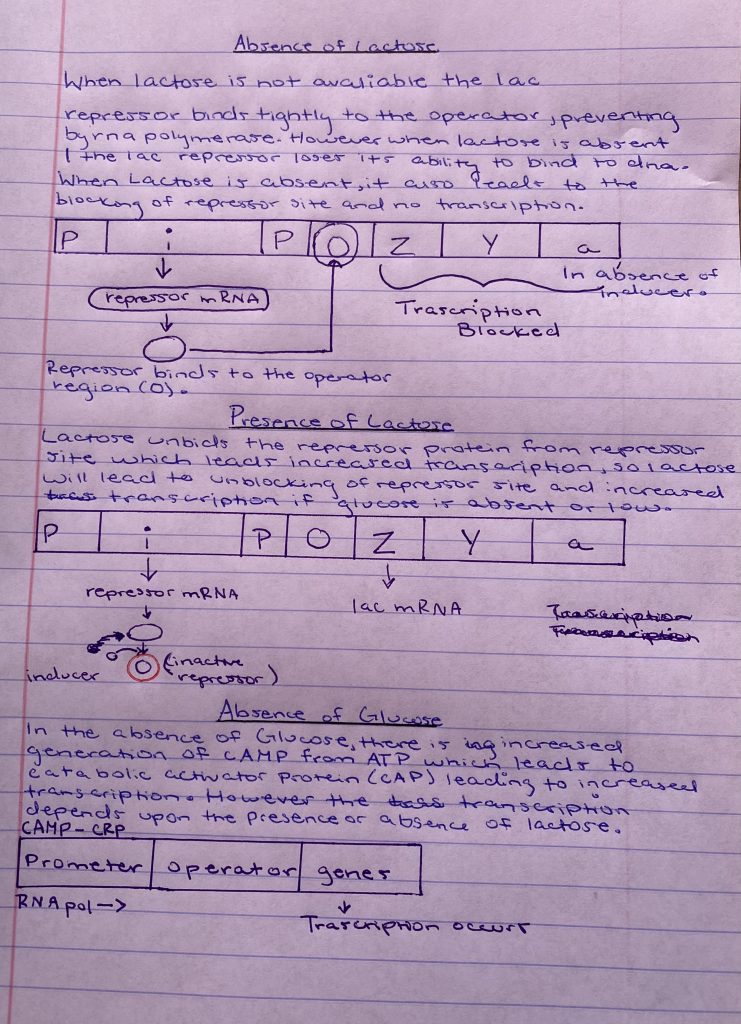
Lac Operon Assignment
Writing assignment 6
Genetics Timeline
Link for timeline: https://prezi.com/view/UvCzILOy4e9kI3GAWNZT/
Genome assignment
Chromosome Maps
Assignment Goal: To use the Internet-based Genes and Disease site (NCBI) to view the assignment of genes to chromosomes.
Assignment: Access the Genes and Disease site at http://www.ncbi.nlm.nih.gov/books/NBK22183/
Under “Contents”, select “Chromosome Map” (at the very bottom).
A karyotype will appear.
Click on a chromosome.
- WHAT CHROMOSOME DID YOU CHOOSE?
- Chromosome 8.
Above the chromosome image you will see the number of genes and base pairs on that particular chromosome.
2 & 3. STATE THE NUMBER OF GENES AND BASE PAIRS ON THE CHROMOSOME YOU CHOSE.
- 1400 genes and 140 million base pairs.
Scan the chromosome map.
4. LIST ONE GENE THAT IS LOCATED ON THIS CHROMOSOME.
- Werner syndrome (WRN)
5. STATE THE NORMAL FUNCTION OF THE GENE YOU LISTED IN #4.
- Slows the growth rate.
6. STATE THE POSSIBLE DISEASE(S) RELATED TO THIS GENE. (This should be possible by clicking on the gene you stated in #4.)
- Heart disease.
GenBank
7. WHAT IS GENBANK? (You can do an Internet search to find this information.)
- Is the NIH genetic sequence database.
Introduction to BLAST
Assignment Goal: To use the Internet-based site BLAST, Basic Local Alignment Search Tool (NCBI), to search for similarities between nucleotide sequences.
Assignment: Access the BLAST site at http://blast.ncbi.nlm.nih.gov/Blast.cgi
Click on “Nucleotide Blast”
Assume that you found this nucleotide sequence when you sequenced a piece of DNA in the laboratory in which you work:
tgtgtgtagg ggggaaggaa tttagctttc acatctctct tatgtttagt tctctgcatg tgcagttaat cctggaactc cggtgctaag gagagactgt tggcccttga aggagagctc ctccctgtgg atgagagaga aggactttac tctttggaat tatctttttg tgttgatgtt atccaccttt tgttactcca
Enter the above sequence (you may copy and paste) into the “Enter Query Sequence” box at the top of the page. Under “Program Selection” near the bottom of the page, choose “somewhat similar sequence (blastn)”
Click the “BLAST” button at the bottom of the page to run the search.
Give some time for the results of your search to show up.
You will be given significant matches for the sequence that you entered.
8. WHAT IS THE TOP SEQUENCE DESCRIPTION MATCH FOR YOUR QUERY SEQUENCE? DO NOT CHOOSE THE PREDICTED SEQUENCE. For this answer, you should give the description listed.
- Homo sapiens CF transmembrane conductance regulator (CFTR), RefSeqGene (LRG_663) on chromosome 7
9. WHAT DOES THE ENCODED PROTEIN DO IN THE BODY? Search the PubMed site at https://pubmed.ncbi.nlm.nih.gov/ to answer this question. Do not give the disease associated with this gene, but the function of the gene itself. CITE THE PAPER YOU USED TO DETERMINE THE PURPOSE OF THE ENCODED PROTEIN.
- “The cystic fibrosis transmembrane conductance regulator (CFTR) protein helps to maintain the balance of salt and water on many surfaces in the body, such as the surface of the lung.”
Basics of the CFTR Protein. (n.d.). Retrieved December 08, 2020, from https://www.cff.org/Research/Research-Into-the-Disease/Restore-CFTR-Function/Basics-of-the-CFTR-Protein/
Click on the top match to find the following.
10. FOR WHAT DISEASE IS A MUTATED FORM OF THIS GENE RESPONSIBLE? You should be able to get this information from the description of the gene.
- Cystic fibrosis
11. ON WHAT CHROMOSOME IS THE GENE LOCATED? You should be able to get this information by clicking on the description of the gene.
- The seventh
12. Scroll to the first described sequence that does not have 100% Query Cover. WHAT ORGANISM IS THE SOURCE OF THIS DNA?
- Humans
13. HOW MANY GAPS OCCUR BETWEEN THE TWO SEQUENCES (THE ONE YOU SUBMITTED AND THE FIRST ONE THAT HAS < 100% QUERY COVER)?
- 0
14. WHAT IS A GAP IN SEQUENCE ALIGNMENTS? (This is something you’ll have to search for elsewhere.)
- “Gaps are the bits that get left behind when you try to align DNA or protein sequences and have to use padding or null characters to match homologous residues.”
Higgins, D. G., Blackshields, G., & Wallace, I. M. (2005). Mind the gaps: progress in progressive alignment. Proceedings of the National Academy of Sciences of the United States of America, 102(30), 10411–10412
BLAST has more sequences than ‘just’ human gene sequences. Also, you can also do BLAST searches using an accession number that has been assigned to a particular sequence when it has been entered into the database. Go back to the Blast home page (http://blast.ncbi.nlm.nih.gov/Blast.cgi ) and again choose “Nucleotide Blast”. Look up the following sequence using the given accession number. (Again, click on the “BLAST” button at the bottom of the page after you have entered the accession number.)
15. STATE WHAT THE FOLLOWING GENE IS: NC_045512.2
(Give the complete description of the gene or gene product.)
- Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome
16. SCROLL DOWN THE LIST BELOW THIS SEQUENCE. THERE ARE MANY SEQUENCES THAT LOOK SIMILAR. CAN YOU DETECT WHAT IS DIFFERENT ABOUT THE OTHER SEQUENCES?
- The numbers on the sides are changing but other than that no.
Introduction to Swiss-Prot to Study Protein Sequences
Assignment Goal: To use the Internet-based site ExPASy (Expert Protein Analysis System) to translate cDNA, and the Internet-based database UniProt KB/Swiss-Prot to access a complete polypeptide.
17. WHAT IS cDNA?
- Is DNA being synthesized from a single stranded RNA (eg., messenger (mRNA)) template in a reaction catalyzed by the enzyme reverse transcriptase.
Assignment: Access the BLAST site at http://blast.ncbi.nlm.nih.gov/Blast.cgi
Click on “Nucleotide Blast”
Enter the following sequence: ACATTTGCTTCTGACACAATTGTGTTCACTAGCAACCTCAAACAGACACCATGGTGCATCTGACTCCTGAGGAGAAGTCTGCCGTTACTGCCCTGTGGGGCAAGGTGAACGTGGATGAAGTTGGTGGTGAGGCCCTGGGCAG
18. USING THE SAME PROGRAM YOU USED IN THE INTRODUCTION TO BLAST ABOVE, WHAT IS THE SEQUENCE MATCH?
- Homo sapiens partial HBB gene for hemoglobin beta chain, exon 1, isolate 04593664
Now access the ExPASt translate tool at https://web.expasy.org/translate/
Enter the above DNA sequence.
Click “Translate Sequence”.
19. HOW MANY 5’ TO 3’ FRAMES DID YOU OBTAIN?
- 3 frames
20. BASED UPON THE LENGTH OF THE POLYPEPTIDE, WHICH FRAME(S) IS (ARE) MOST LIKELY THE CORRECT ONE?
- Frame 2
Amino Acid Sequence Comparisons
Assignment Goal: To use the Internet-based site ExPASy program SIM to align two amino acid sequences. Knowing the sources of these sequences will allow one to determine the mutation and potential cause of a human disease.
Assignment: Access the ExPASy site at https://web.expasy.org/sim/
Copy and paste each of the following sequences into the “Sequence” text boxes in SIM.
Person 1/Sequence 1:
MGAPACALALCVAVAIVAGASSESLGTEQRVVGRAAEVPGPEPGQQEQLVFGSGDAVELSCPPPGGGPMGPTVWVKDGTGLVPSERVLVGPQRLQVLNASHEDSGAYSCRQRLTQRVLCHFSVRVTDAPSSGDDEDGEDEAEDTGVDTGAPYWTRPERMDKKLLAVPAANTVRFRCPAAGNPTPSISWLKNGREFRGEHRIGGIKLRHQQWSLVMESVVPSDRGNYTCVVENKFGSIRQTYTLDVLERSPHRPILQAGLPANQTAVLGSDVEFHCKVYSDAQPHIQWLKHVEVNGSKVGPDGTPYVTVLKTAGANTTDKELEVLSLHNVTFEDAGEYTCLAGNSIGFSHHSAWLVVLPAEEELVEADEAGSVYAGILSYGVGFFLFILVVAAVTLCRLRSPPKKGLGSPTVHKISRFPLKRQVSLESNASMSSNTPLVRIARLSSGEGPTLANVSELELPADPKWELSRARLTLGKPLGEGCFGQVVMAEAIGIDKDRAAKPVTVAVKMLKDDATDKDLSDLVSEMEMMKMIGKHKNIINLLGACTQGGPLYVLVEYAAKGNLREFLRARRPPGLDYSFDTCKPPEEQLTFKDLVSCAYQVARGMEYLASQKCIHRDLAARNVLVTEDNVMKIADFGLARDVHNLDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLLWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTHDLYMIMRECWHAAPSQRPTFKQLVEDLDRVLTVTSTDEYLDLSAPFEQYSPGGQDTPSSSSGDDSVFAHDLLPPAPPSSGGSRT
Person 2/Sequence 2:
MGAPACALALCVAVAIVAGASSESLGTEQRVVGRAAEVPGPEPGQQEQLVFGSGDAVELSCPPPGGGPMGPTVWVKDGTGLVPSERVLVGPQRLQVLNASHEDSGAYSCRQRLTQRVLCHFSVRVTDAPSSGDDEDGEDEAEDTGVDTGAPYWTRPERMDKKLLAVPAANTVRFRCPAAGNPTPSISWLKNGREFRGEHRIGGIKLRHQQWSLVMESVVPSDRGNYTCVVENKFGSIRQTYTLDVLERSPHRPILQAGLPANQTAVLGSDVEFHCKVYSDAQPHIQWLKHVEVNGSKVGPDGTPYVTVLKTAGANTTDKELEVLSLHNVTFEDAGEYTCLAGNSIGFSHHSAWLVVLPAEEELVEADEAGSVYAGILSYRVGFFLFILVVAAVTLCRLRSPPKKGLGSPTVHKISRFPLKRQVSLESNASMSSNTPLVRIARLSSGEGPTLANVSELELPADPKWELSRARLTLGKPLGEGCFGQVVMAEAIGIDKDRAAKPVTVAVKMLKDDATDKDLSDLVSEMEMMKMIGKHKNIINLL
GACTQGGPLYVLVEYAAKGNLREFLRARRPPGLDYSFDTCKPPEEQLTFKDLVSCAYQVARGMEYLASQKCIHRDLAARNVLVTEDNVMKIADFGLARDVHNLDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLLWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTHDLYMIMRECWHAAPSQRPTFKQLVEDLDRVLTVTSTDEYLDLSAPFEQYSPGGQDTPSSSSSGDDSVFAHDLLPPAPPSSGGSRT
Click the “User entered sequence” button for each sequence entered.
Submit the sequences for comparison.
21. DO YOU SEE ANY DIFFERENCES BETWEEN THE TWO AMINO ACID SEQUENCES? (Look for the absence of an asterisk, which indicates the same amino acid in both sequences.)
- Yes, in 781 & 361
22. WHY WOULD DIFFERENCES BE IMPORTANT AND POSSIBLY CLINICALLY USEFUL?
Return to the BLAST home page (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Run a PROTEIN BLAST search to identify the polypeptide which you have been analyzing. (You may use either sequence.)
- Differences can lead to a different amino acid forming that can also lead to the changing of the function of a gene
23. WHAT IS THE PROTEIN THAT YOU WERE ANALYZING?
Click on “Description”, then choose “Gene”.
- Fibroblast growth factor receptor 3 isoform 1 precursor [Homo sapiens]
24-25. REFLECT ON ONE THING YOU ALREADY KNEW, AND ONE THING THAT YOU LEARNED FROM DOING THIS ASSIGNMENT.
- I already knew about CF being Cystic Fibrosis and also that the WRN gene that can lead to heart disease. Since Werner syndrome is the early development of various types of cancer commonly known as hardening of the arteries, which can lead to a heart attack. Something that I learned from doing this assignment is “WHAT IS A GAP IN SEQUENCE ALIGNMENTS?” I never knew that it was bits left when you try to align DNA or protein sequences. That is very interesting to know.
Romanov Assignment
History
You may use various Internet sources here, but please cite any sources that you use (unless I have given it to you). You will be using the following source for another section in this assignment, but it might help you answer some of the questions in this section. If you use this source to answer the following questions, you do not need to cite it. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0004838 (You should be able to access the entire article. You may need to copy and paste the site address.)
1. Who were the Romanov’s (in Russian history)?
- the last Russian monarchy.
2. How long were the Romanovs in power in Russia?
- For over 300 years, the Romanovs ruled the country of Russia.
3. Nicholas II was the last Romanov to hold power in Russia. What was his title?
- Russian Tsar
4. Politically, what happened to Nicholas II?
- abdicated his crown in favor of his brother Grand Duke Michael, who declined to accept the throne.
5. Who took control after Nicholas II abdicated the throne?
- A provisional government took control after Nicholas II abdicated the throne, and then the 1917 Revolution led to the Bolshevik, communist uprising and Vladimir Lenin taking control.
6. What happened to Nicholas II and his family after he abdicated the throne?
- Nicholas and his family – his wife, the Tsarina Alexandra, and their five children: Olga, Tatiana, Maria, Anastasia, and the Tsarevich (Crown Prince) Alexei were held in exile in Yekaterinburg, Russia.
7. One of the reasons that the family of Nicholas II was executed was because there was a fear that the White Russian Army would save them. Who was the White Russian Army?
- The White armies were anti-Bolshevik forces who participated in the Russian Civil War.
Hemophilia
The pedigree chart below comes from the Module powerpoint lecture notes.
8. How was Nicholas II wife, Alix, related to Queen Victoria of England?
- That’s her grandmother.
Both Queen Victoria and Alix are designated as being carriers for hemophilia.
9. In a couple of sentences, describe the physiology of the disease hemophilia. (Yes, I know it is severe bleeding because the blood cannot clot. But WHY can’t the blood clot?)
- When you bleed, your body normally pools blood cells together to form a clot to stop the bleeding. The clotting process is encouraged by certain blood particles. Hemophilia occurs when you lack sufficient blood-clotting proteins (clotting factors). There are several types of hemophilia, and most forms are inherited.
10. What does it mean to be a carrier for a disease?
To be a carrier is to have the capability to pass on a genetic mutation associated with a disease that may or may not be displayed through symptoms. To be more specific, a carrier has one allele for the certain condition but that allele does not express the phenotype.
Use the following source for the questions 8 & 10: http://www.ncbi.nlm.nih.gov/pubmed/20557352 (You won’t be able to access the entire article, but the abstract will give you the information you need to answer the questions.)
11. What type of hemophilia (A or B) is (probably) represented in the pedigree chart?
Hemophilia b
12. Using your knowledge from Module 4, on what chromosome is the gene that, when mutated, causes hemophilia?
X chromosome
13. Describe the mutation that apparently caused hemophilia in Alix, (and probably all of the European families that had hemophilia). Be very specific.
A substitution mutation on exon 4, gene f9. This led to a change in amino acid sequence.
14. Using your knowledge from Module 7, describe how the mutation you described in #10 could result in a faulty gene product. Be very specific in your description.
Since there was a substitution, the amino acid sequence was changed leading to a whole new protein.
15. The Romanov’s son, Alexis, had hemophilia. Describe how Alexis genetically acquired hemophilia. (Use a Punnett square. You can either draw a table or line up the genotypes.)
| X | Y | |
| X’ | X’X | X’Y |
| X | XX | XY |
16. Using a Punnett square (again, draw a table or line up the genotypes), explain why only males in the pedigree chart have hemophilia. (Choose at least one of the males represented in the pedigree chart, and show his parents in the Punnett square.)
| X’ | X | |
| X’ | X’X’ | X’X |
| Y | X’Y | XY |
When both parents are just carriers, 75% of the offspring have the gene for hemophilia, 50% of them are carriers and the other 25% has the disease. When males have the trait, since they only have one X chromosome, they are affected and develop the disease unlike females that have 2 X chromosomes. %
17. Is it possible for a female to inherit hemophilia, and, if so, how?
Females can have hemophilia but a much smaller chance. Either both X chromosomes are affected or a female that is a carrier can show mild symptoms of hemophilia.
18. Using a Punnett square (again, draw a table or line up the genotypes), what is the probability the daughter of a mother who is a carrier and a father who does not have the disease, will be a carrier?
| X | Y | |
| X’ | X’X | X’Y |
| X | XX | XY |
The probability that a daughter will become a carrier is 25%
19. Using a Punnett square (again, draw a table or line up the genotypes), what is the probability that 4 daughters of a mother who is a carrier and a father who does not have the disease, will be a carrier?
The probability that 4 daughters of a carrier mother and a normal father will be carriers is 50%, so theoretically, two daughters have both wild-type alleles (H) while the other two are carriers (one H and one h allele). Therefore, there is no way that any daughter could be affected by hemophilia. On the other hand, the probability that a son will be affected by the disease is 50% since he only inherits one X chromosome from the mother.
| XH | Y | |
| XH | XHXH | XHY |
| Xh | XHXh | XhY |
20. Some historians speculate that Alexei’s hemophilia condition could have led to the Russian Revolution. Explain.
The Romanovs, particularly Tsar Nicholas II, believed that the spiritual healer Rasputin could treat Alexei’s hemophilia. Because of Rasputin’s role, he developed a close relationship with the Romanov family which was heavily criticized by the Russian people due to Rasputin’s reputation as a fraud. When Nicholas II left St. Petersburg to fight in World War I, Rasputin remained in the palace, which increased his sphere of influence within the Russian government. Therefore, while “healing” Alexei, Rasputin took advantage of his position which exposed the weaknesses of the Romanov family. Because of the vulnerability of the Romanov family (along with economic and political instability), it was the perfect opportunity for the Bolsheviks to start a revolution.
Molecular Analysis of People in a Mass Grave
Use the following source to help you answer the following questions: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0004838 (You should be able to access the entire article.)
21. Two “graves” were discovered near Yekaterinburg, Russia. Describe the number of bodies in each grave.
At the two “graves” near Yekaterinburg, Russia, nine members were buried in one mass grave and two children were buried in the other grave.
22. When were these graves discovered?
The larger mass grave was discovered in 1991 and the smaller grave was discovered in 2007. The smaller grave was located about 70 meters away from the larger grave.
23. What type of testing was done to confirm sex and familial relationships among the remains found in the mass grave?
Forensic DNA testing was performed on the bodies using mitochondrial DNA. These methods include autosomal short tandem repeat analysis (STR) and Y-STR testing.
24. Genetically, what does STR “stand” for? Be very specific in your answer.
Autosomal Short Tandem Repeat analysis is a technique used to examine the differences between two different samples of DNA in the same locus. Certain DNA loci are extracted, amplified with PCR, and run under gel electrophoresis to determine how many repeats of the STR sequence exist. Differences in STR repeats can be seen from one sample to another.
25. Mitochondrial DNA testing was also done on both Nicholas II and Alix. Why was information from Alix’s, but not Nicholas’, mitochondrial DNA used to identify three females as belonging to Alix?
When looking at mitochondrial DNA, the offspring inherit mitochondrial DNA from the mother. Therefore, when identifying the three females as the daughters of Alexandra, mitochondrial DNA can be used to find similarities between Alexandra and her children.
26. HRH Prince Philip, the Duke of Edinburgh, provided mitochondrial DNA used to identify Alix and her three daughters. HRH Prince Philip, the Duke of Edinburgh, is married to Queen Elizabeth II of England. Wait, isn’t Queen Elizabeth II related to Queen Victoria? So why was Prince Philip’s mitochondrial DNA used?
It turns out that Prince Philip is also a descendant of Queen Victoria, so he inherited her mitochondrial DNA and could provide samples to identify Alexandra and her three daughters. Based on their family tree, Prince Philip and Queen Elizabeth II are third cousins through Queen Victoria.
27. Who was missing from the mass grave?
Before the discovery of the second grave in 2007, there were two children missing from the first mass grave: Alexei and one of the daughters (either Anastasia or Maria).
28. The Duke of Fife and Princess Xenia provided mitochondrial DNA used to identify Nicholas. One of these is a female and another is a male. Does that matter? What general statement can you make about their genetic
relationship to Nicholas and Alexandra?
This matters because without Xeina’s DNA, it wouldn’t be possible to match the mitochondria because it’s passed from mother to child. For this reason, her’s would be necessary to match with Nicholas.
29. What was discovered in the mitochondrial DNA of Nicholas that was not identified in either the Duke of Fife or Princess Xenia?
There was a single point heteroplasmy at position 16169, whereas both the Duke and Xeina were fixed at position 16169.
30. What is the term given to the existence of two (or more) genetically different mitochondria in the cell?
Heteroplasmy is when there are two or more genetically different mitochondria in the cell.
31. What three types of DNA were used to test the remains found in a second grave?
Ancient DNA, mitochondrial DNA, and nuclear DNA
32. Of the three types of DNA you listed in #31, which one would have been used specifically to identify Alexis?
Mitochondrial DNA
33. What was the source of the DNA used to identify Alexis?
Her teeth and other bone fragments
34. Was Anastasia in the grave in which Alexis was found?
No, she was not
Who Wants to Be Anastasia?
Apparently, about 200 people have wanted to be Anastasia and have claimed to be her! One of the most famous imposters was a woman named Anna Anderson (Manahan).
Refer to the following source to help you answer some of the following questions: http://www.nature.com.proxy.lib.odu.edu/ng/journal/v9/n1/pdf/ng0195-9.pdf (Please note that this is a PDF of the article and is provided via the ODU Library.)
35. Give a brief history (2-3 sentences) of Anna Anderson-both her claims and what is thought to be true.
Anna Anderson laid her claim to be the Royal Duchess Anastasia in the 1920s. However, those who doubted her claimed that she was Franzisca Schanzkowska who was admitted to a mental hospital which she escaped in 1920. DNA testing was performed on samples from Anderson and compared to the Tsarina and one of Schanzkowska’s great nephews. The results determined that she could not be maternally related to the Tsarina but did share DNA with the great nephew of Schanzkowska, meaning Anderson and Schanzkowska were on in the same.
36. Where in the US did Anna Anderson eventually settle?
Charlottesville, Virginia
37. Whom did she eventually marry?
John Manahan (Mintz, 1983)
38. What were the sources of Anna Andersons’s nuclear DNA?
Small bowl and hair samples
39. What were the sources of Nicholas’ and Alix’s nuclear DNA?
Blood sample from a known relative of the Tsarina
40. What type of analysis was done on DNA from Anna Anderson, Nicholas, and Alix?
mtDNA and STR analysis
41. Anna Anderson’s mitochondrial DNA was compared to the mitochondrial DNA of what two people?
Carl Maucher and Duke of Edinburgh
42. A hypervariable region of the mitochondrial DNA was analyzed. Define a hypervariable region?
Hypervariable region- region of DNA which is highly susceptible to change
43. What were the conclusions from the mitochondrial DNA comparisons?
The conclusions discussed in the article were that Anderson was not Anastasia nor was she at all related to the royal family. Anderson was in fact related to the great nephew of Schanzkowska and was most likely Franzisca Schanzkowska, who escaped the mental hospital in the 1920s
44. The article which describes the analysis of Anna Anderson’s DNA was published in 1995. When were all of Nicholas’ and Alix’s children finally accounted for?
2007 (Coble et al., 2009)
45. What did you learn from doing this assignment? (Each person in a group should answer this question. It is not a group answer.)
Batonya: The whole story of the Romanov family was confusing to me. However, I learned who the Romanov were, and the importance of their family. I had the chance to relearn one thing from the Romanov family story. Which was, “Who took control after Nicholas II abdicated the throne?” I forgot that Vladimir Lenin took control after Nicholas II abdicated the throne. It was fun to get the chance to relearn of what happened to the Romanov family.
Reagan: From this assignment, I learned that there was a lot of confusion and mystery surrounding the death of the Romanov family. As the last royal family, the Romanovs symbolized the end of imperial rule and the beginning of communist rule, so many efforts were made to erase the Romanov family from history.
Tyler: Although I already knew the story of the Romanov family, I had no idea there was this much work put into genetically uncovering and finding them. I also learned more about the ways DNA can be used to identify corpses.
Luwam: I learned about the importance of pedigree charts and the patterns of how diseases can be passed down through families, even if the parents are unaffected. This assignment makes me want to go through my own family history too.
Melissa: Prior to this assignment I was familiar with the Romanov family, but I was unaware of how much genetic detective work went into the discovery of the missing princess. I did not know that the princess had an imposter who moved to the United States and claimed it as her home. Learning that piece of information, especially since her final place of residence is not too far from Norfolk, is incredibly interesting.
Are you still interested in the life of the last Tsar of Russia and his relationship to British royalty? The headline for the following article showed up on my Internet browser earlier this year. While I can’t vouch for it as it did not appear in a peer-reviewed journal, it might be interesting reading for you.
BY PLACING MY NAME BELOW (AND INCLUDING A DATE), I HAVE REVIEWED THE ANSWERS ON THIS ASSIGNMENT, AND AGREE TO THEM.
Luwam Abeselom 12/11/20
Batonya Southerland 12/11/20
Reagan Richardson 12/11/20
Melissa Leigh 12/11/20
Tyler Filippini 12/11/20
References:
1. Mintz J. 1983. Woman Claiming Kinship to Czar Disappears in Va. The Washington Post. WP Company.
2. Coble MD, Loreille OM, Wadhams MJ, Edson SM, Maynard K, Meyer CE, Niederstätter H, Berger C, Berger B, Falsetti AB, Gill P, Parson W, Finelli LN. 2009. Mystery solved: the identification of the two missing Romanov children using DNA analysis. PloS one. Public Library of Science.
3. Carrier. (n.d.). Retrieved December 01, 2020, from https://www.genome.gov/genetics-glossary/Carrier