Abstract: Ticks not only feed on blood, but are one of the known vectors of pathogens in the environment. When a bloodmeal analysis is performed, scientists are capable of identifying sources of blood that remain in their digestive tract upon their death. By knowing more about the relationship between the ticks and their vertebrate of their choosing, the capacity to understand how diseases may be transmitted is greatly increased. The purpose of this research project was to identify the DNA of the remaining bloodmeal from the tick, also known as the tick’s food source, to gain a better understanding of the variety of vertebrates the ticks feed on, which may give insights on transmission of disease. Gradient polymerase chain reaction was performed on 12 samples of tick DNA from the Hoffler Creek Wildlife Preserve in Portsmouth, VA; during this step samples 1-8 were lost. A simple DNA extraction was then performed on samples 9-12 by liquifying the bloodmeal using Digestion Buffer and proteinase K. Post incubation, RNase A and lysis buffer were used to further break down the nuclear envelope. The samples were then purified by using column based purification efforts where the sample was treated with two wash buffers and an elution buffer. Afterwards, Sanger dideoxy sequencing was performed and the blast results were obtained.
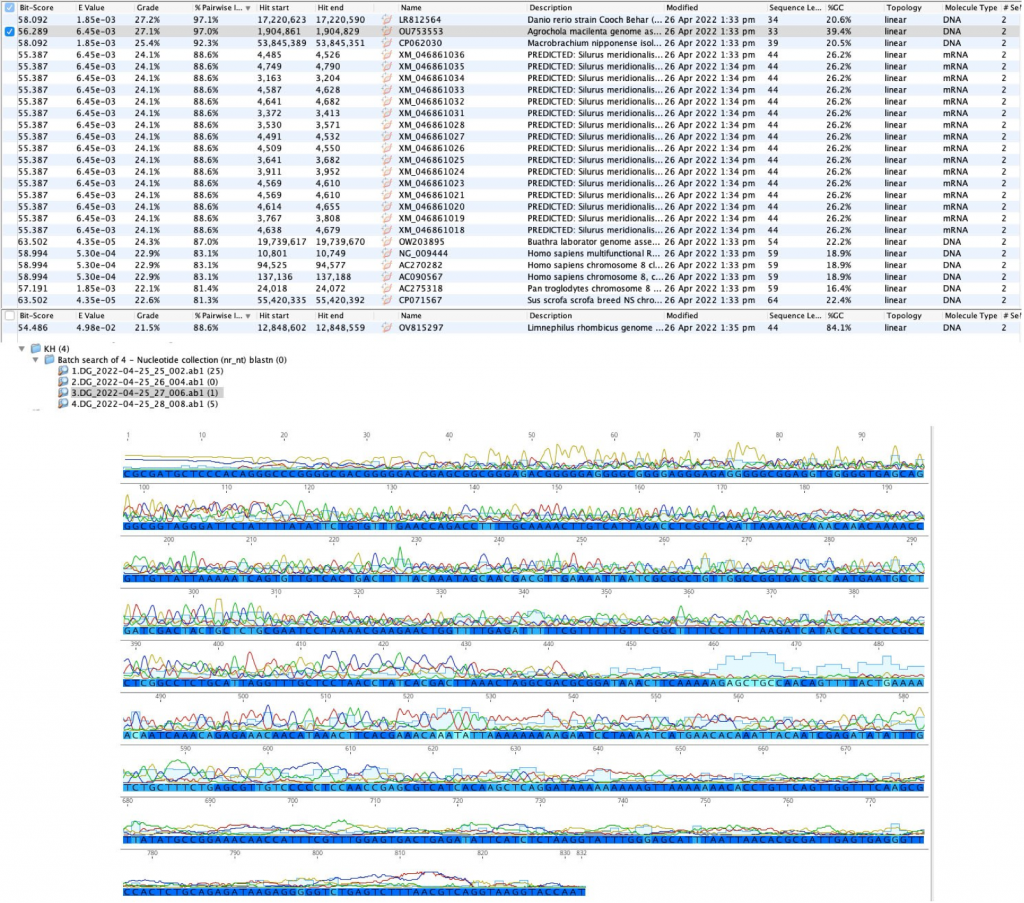
Conclusions: Given that none of the DNA samples were remotely identified as a tick, the results reflect poor sequencing. It appears as though there were several random hits. Also, the purity of the sample given the peaks present on the sequence in the photo above shows purifications attempts were not successful. An improvement that could have been made prior to DNA sequencing would be to ensure that the strip tube was sealed properly. Additionally, spectrophotometry should have been performed to ensure that the sample was pure, if not further purification could have been performed. Attempting the experiment with different primer sets could have also increased the success of the experiment.