⭐️Writing #1⭐️

⭐️Writing #2⭐️
Mia Littlejohn
Ms. Rinehart-Kim
BIOL 294
1/29/24
Identification of a Primary Article versus a Review Article
A primary article or source denotes a firsthand topic or event recorded originally by a participant in, a witness to, or an author of that topic or event. More importantly, these sources provide an account of events as they happened originally. Primary sources can include an eyewitness account, book, statistical manuscript, or an article (Academic Guides at Walden University). In research, primary articles proffer information about the findings of a novel research. Researchers who conduct firsthand research experimentation write primary articles.
A review article differs entirely from a primary article on several accounts. In particular, it is a secondary information source. As a secondary information source, a review article examines previously published research. As such, it is also denoted as a literature review. More importantly, a review article does not provide novel results of an experiment(Academic Guides at Walden University). Rather, it offers insights into the current thinking about a specific subject or research.
Scientific peer review is a standardized process wherein multiple scholarly experts scrutinize the ideas, research, or scholarly work of another author. This process promotes the publication of first-rate scholarly work or research in reputable journals. It also checks the validity and authenticity of primary research findings. This process begins after a scientist submits a manuscript to a journal detailing the conclusions, research design, purpose, and results of an original study (Kelly et al. 229). The journal editors review the manuscript to verify the alignment of the journal and the paper’s subject matter. This is the initial assessment, which only a few papers pass. Following a rigorous credibility test, the editors submit the manuscript to reputable researchers (reviewers) who perform formal peer review (Kelly et al. 229). The reviewers examine all sections of the manuscript to ascertain the veracity of the research. They also identify errors and provide definitive recommendations to the editors on whether the manuscript is genuine or should not be published.
The article written by Huang and colleagues is a review article. This is because it uses statements informing readers that their research is a review of original research that was published previously. In the abstract, for instance, the authors claim that their study reviews other published studies focusing on CRISPR systems (Huang et al. 5934). The term – reviews – indicates that this is not original research. Comparatively, the article written by Tsou and colleagues is a primary source. The authors combined CRISPR and reverse transcription-isothermal recombinase polymerase amplification to identify SARS-CoV-2 (Tsou et al. 1). The authors made a firsthand investigation to achieve a particular goal, thereby making their study an original research.
Works Cited
Academic Guides at Walden University. “Evaluating Resources: Primary & Secondary Sources.” Academic Guides at Walden University, https://academicguides.waldenu.edu/library/evaluating/sources
Huang, Yueying, et al. “Development of Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated Technology for Potential Clinical Applications.” World Journal of Clinical Cases, vol. 10, no. 18, June 2022, pp. 5934–45. https://doi.org/10.12998/wjcc.v10.i18.5934.
Kelly, Jacalyn, et al. “Peer Review in Scientific Publications: Benefits, Critiques, & A Survival Guide.” EJIFCC vol. 25,3 227-43. 24 Oct. 2014, pp. 227–243. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4975196/pdf/ejifcc-25-227.pdf
Tsou, Jen-Hui, et al. “Rapid and Sensitive Detection of SARS-CoV-2 Using Clustered Regularly Interspaced Short Palindromic Repeats.” Biomedicines, vol. 9, no. 3, Feb. 2021, p. 239. https://doi.org/10.3390/biomedicines9030239.
⭐️Writing #3⭐️
1.E Burrow, Patricias ;Gonzalez-Garay ML;Rasmussen JC;Aldrich MB;Guilliod R;Maus EA;Fife CE;Kwon S;Lapinski PE;King PD;Sevick-Muraca EM;, P. Lymphatic abnormalities are associated with RASA1 gene mutations in mouse and man. Proceedings of the National Academy of Sciences of the United States of America (2013). Available at: https://pubmed.ncbi.nlm.nih.gov/23650393/. (Accessed: 19th February 2024)
⭐️Writing #4⭐️
The article I selected for this assignment studies how the RASA1 gene is associated with lymphatic abnormalities in both a mouse and a man. The RASA1 gene is most commonly known for capillary malformation however, its role in lymphatic abnormalities have yet to be studied deeply. To gather more information on this subject the authors of the article put a PKWS patient to the test with some infrared lymphatic imaging. With this testing, researchers discovered that abnormal lymphatic vasculature was evident in both the infected and uninfected legs of the. PKWS patient. As for the mice tested in this experiment with a RASA1 deficiency, the findings of dermal lymphatic hyperplasia and dilated vessels were evident. From the results gathered in this experiment researchers concluded that the RASA1 gene mutations were responsible for aberrant lymphatic architecture and functional abnormalities
1.E Burrow, Patricias ;Gonzalez-Garay ML;Rasmussen JC;Aldrich MB;Guilliod R;Maus EA;Fife CE;Kwon S;Lapinski PE;King PD;Sevick-Muraca EM;, P. Lymphatic abnormalities are associated with RASA1 gene mutations in mouse and man. Proceedings of the National Academy of Sciences of the United States of America (2013). Available at: https://pubmed.ncbi.nlm.nih.gov/23650393/. (Accessed: 19th February 2024)
⭐️Writing #5⭐️
The Evolutionary Origin of Multiple Sclerosis: Linking Ancient DNA and Genetic Risk in European Populations
Mia Littlejohn
Old Dominion University
Ms. Rinehart-Kim
BIOL 294
04-08-24
The Evolutionary Origin of Multiple Sclerosis: Linking Ancient DNA and Genetic Risk in European Populations
Ancient DNA discoveries have signaled some fascinating interconnections between past migrations and contemporary health conditions. In a recent article published on January 10, 2024, in The Washington Post titled “Ancient DNA helps trace multiple sclerosis origins in European descendants” by Carolyn Y. Johnson, it reveals that a complex study conducted uncovered the roots of multiple sclerosis (MS) among Europeans. This paper will discuss the findings of this research and how they align with a scientific review by Hernández-Preciado et al. (2024) on gene expression during pregnancy among females with multiple sclerosis.
Summary of the Newspaper Article
About 5,000 years ago, there were migrations from Asia to Europe, bringing immunity-related genetic variants. These ancient immigrants have now been shown to be associated with higher genetic risks of developing multiple sclerosis among Europeans (Johnson, 2024). As the newspaper article mentioned, this migration brought about genetic adaptations that conferred an advantage against ancient infections. It then raises questions about whether these alleles might explain why people of Northern European descent tend to develop multiple sclerosis at such high rates.
The research explained in this Washington Post article unravels a long-standing mystery in medical science: why are individuals of Northern European descent most vulnerable to MS? It suggests an evolutionary trade-off, which means multiple sclerosis could be linked to immune system genes (Johnson, 2024). These gene variants were once favored by natural selection because they provided adequate protection against infections and parasites borne by livestock thousands of years ago. However, they now contribute to a higher susceptibility to MS among their descendants.
Supporting Evidence
Gene expression during pregnancy in mothers with multiple sclerosis was recently investigated in a scientific review done by Hernandez-Preciado et al. (2024). The group found that 42 genes were differentially expressed during pregnancy and had already been linked to the disease, including IL-10 signaling, IL-17 signaling antigen processing and presentation, and Th17 cell differentiation, which point to how complicated immune molecular profiles like those seen in MS have come about. Consequently, modern mutations that are advantageous to immunity can also be associated with multiple sclerosis.
Therefore, the author argues for prehistoric genetic adaptations that make contemporary Europeans susceptible to multiple sclerosis (Hernández-Preciado et al., 2024). Essentially, this knowledge helps an individual know how best to take care of a woman’s health after she has conceived so as not to develop extra complications related to MS. This argument is important because it emphasizes genetics’ historical importance and relevance concerning multiple sclerosis.
Alignment of Findings
The findings made by Hernández-Preciado et al. (2024) indicate that the information presented in the newspaper article by Johnson (2024) is accurate. A review of both articles shows that the findings made by both authors are consistent. The Washington Post accurately depicts the main points of the study, highlighting how some immune genes that were advantageous during evolution act as risk factors for multiple sclerosis (Johnson, 2024). On the other hand, the scientific article explores individual genes and pathways implicated in pregnancy-related MS, further supporting the genetic complexity of the disease. This article vindicates the popular press. As a result, it makes it more credible and hence reliable. In the review, it is apparent that The Washington Post’s research successfully connected ancient migration patterns and subsequent genetic adaptive changes to modern prevalence rates of multiple sclerosis among populations with European ancestry. This data helps to identify where this disease came from and points out areas for further inquiry and treatment.
References
Hernández-Preciado, M. R., Torres-Mendoza, B. M., Mireles-RamÃrez, M. A., Kobayashi-Gutiérrez, A., Sánchez-Rosales, N. A., Vázquez-Valls, E., & Marquez-Pedroza, J. (2024). Gene expression in multiple sclerosis during pregnancy based on integrated bioinformatics analysis. Multiple Sclerosis and Related Disorders, 82, 105373.
Johnson, C. Y. (2024, January 10). Ancient DNA helps trace multiple sclerosis origins in European descendants. Washington Post; The Washington Post. https://www.washingtonpost.com/science/2024/01/10/ancient-dna-multiple-sclerosis-european-ancestry/
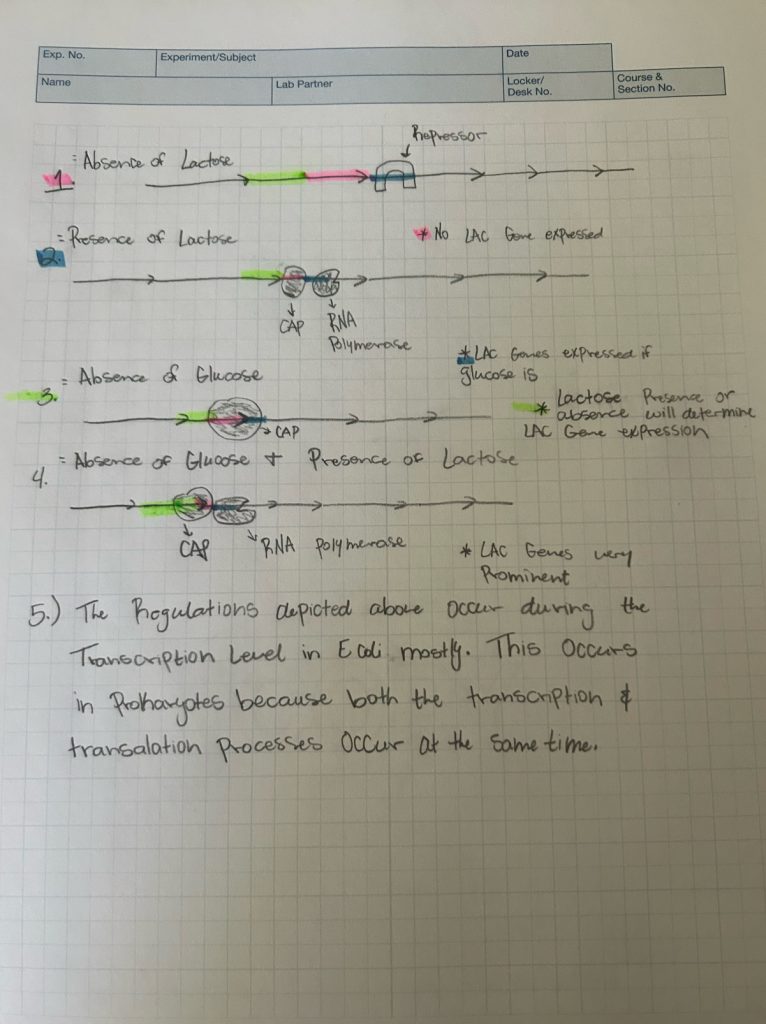
⭐️Lac Operon Assignment⭐️
⭐️Genome Assignment⭐️
- Chromosome #11
- 2,000 genes contained
- over 130 million base pairs
- Multiple Endocrine Neoplasia
- Releases hormones into the bloodstream
- Homo sapiens CFTR (CFTR) gene, partial cds
- Encodes a member of the ABC
- Cystic Fibrosis
- Chromosome #7
- Camelus ferus
- Bactrian camel
- yes, because the species is a form of bacteria
- sapajus apella, monkey (tufted capuchin), yes
- 1.24%
- protein import into nucleus
- encodes ubiquitin-like protein
- encodes multifunctional protein
- Makes a protein that encodes copper and zinc ions
- ALS, Lou Gehrig’s disease
- NIH genetic sequence database
- complementary DNA
- MVHLTPEEKSAVTALWGKVNVDEVGGEMVHLTPEEKSAVTALWGKVNVDEVGGEALMVHLTPEEKSAVTALWGKVNVDEVGGEAL
- Sequence 2 has a space sequence one does not
- The space in sequence 2
- yes, in sequence 2
- Fibroblast
- encodes a member of the FGFR family
- Cervix cancer
- National Institutes of Health, National Library of Medicine, National Center of Biotechnology Information, and U.S. Department of Health and Human services. The U.S. government oversees these companies
- One positive thing I encountered while doing this assignment is the directions for the assignment are generally very simple and easy to follow, also during this experiment I feel like I learned a lot about the topic in general. One negative thing I encountered during this experiment was that I did have trouble finding the answers to a few of the questions on the website just because there was so much information to look through. Overall I had a great experience with this genome assignment and I learned a lot during the process.
Other Sources Used: