Diffuse low-grade gliomas (LGG) have been reclassified based on molecular mutations, which require
invasive tumor tissue sampling. Tissue sampling by biopsy may be limited by sampling error, whereas
non-invasive imaging can evaluate the entirety of a tumor. This study presents a non-invasive analysis of
low-grade gliomas using imaging features based on the updated classification. We introduce molecular
(MGMT methylation, IDH mutation, 1p/19q co-deletion, ATRX mutation, and TERT mutations) prediction
methods of low-grade gliomas with imaging. Imaging features are extracted from magnetic resonance
imaging data and include texture features, fractal and multi-resolution fractal texture features, and
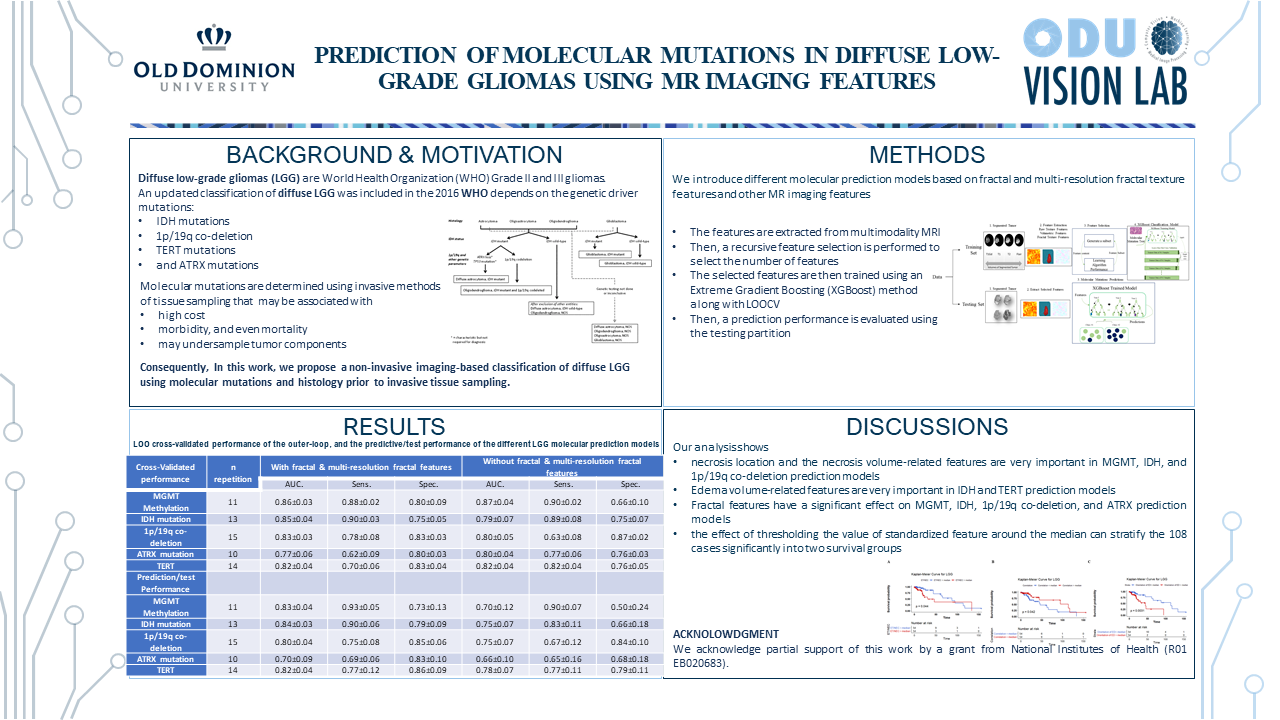
volumetric features. Training models include nested leave-one-out cross-validation to select features,
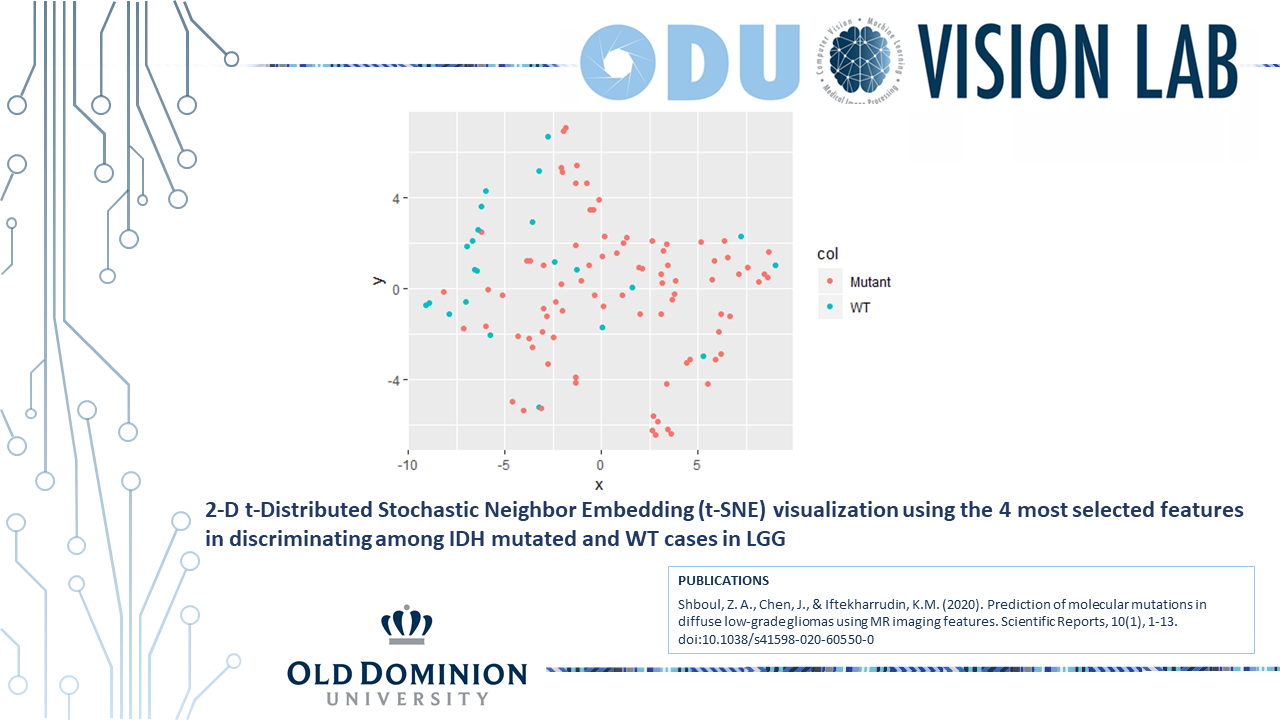
train the model, and estimate model performance. The prediction models of MGMT methylation, IDH
mutations, 1p/19q co-deletion, ATRX mutation, and TERT mutations achieve a test performance AUC of
0.83±0.04, 0.84±0.03, 0.80±0.04, 0.70±0.09, and 0.82±0.04, respectively. Furthermore, our analysis
shows that the fractal features have a significant effect on the predictive performance of MGMT
methylation IDH mutations, 1p/19q co-deletion, and ATRX mutations. The performance of our
prediction methods indicates the potential of correlating computed imaging features with LGG
molecular mutations types and identifies candidates that may be considered potential predictive
biomarkers of LGG molecular classification.
Vision Lab
Computational Intelligence and Machine Vision Laboratory